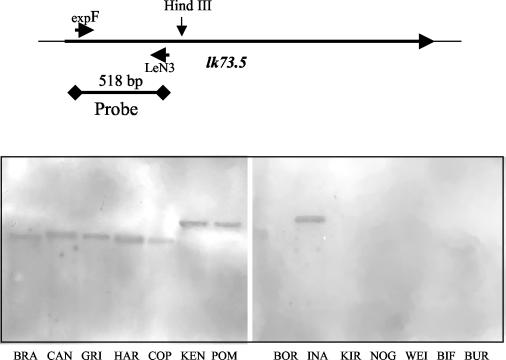
FIG. 6.
Southern blot analysis of genomic DNAs from selected strains of Leptospira spp. DNA of B. burgdorferi, the genomic DNA of which has no sequence homologous to lk73.5, was used as a negative control. Genomic DNAs were digested with HindIII and probed with a biotin-labeled fragment of the 5′ terminus of lk73.5, which has less than 40% overall homology with other leptospiral sphingomyelinase genes. Lanes: BRA, L. interrogans serovar Bratislava; CAN, L. interrogans serovar Canicola; GRI, L. interrogans serovar Grippotyphosa; HAR, L. interrogans serovar Hardjo; COP, L. interrogans serovar Copenhageni; KEN, L. interrogans serovar Pomona type kennewicki; POM, L. interrogans serovar Pomona; BOR, L. borgpetersenii; INA, L. inadai; KIR, L. kirschneri; NOG, L. noguchi; WEI, L. weilii; BIF, L. biflexa; BUR, B. burgdorferi. The schematic on the top shows the positions of the single HindIII site and of the sequence used as a probe.