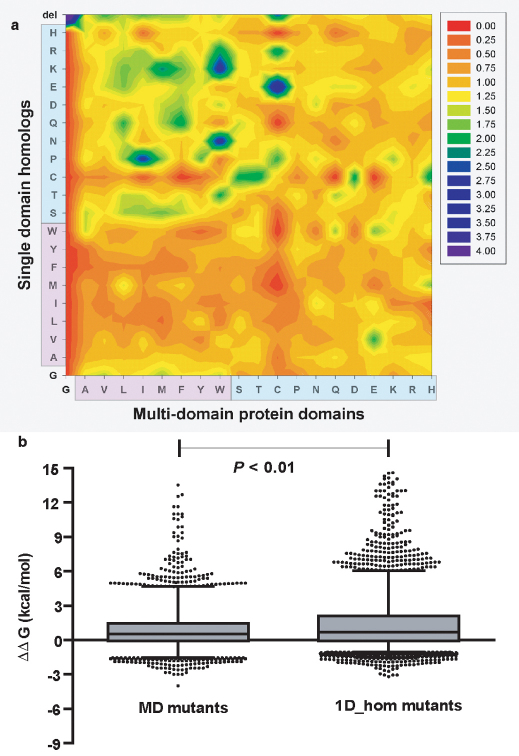
Figure 5. Substitution preferences in single-domain homologues of multi-domain protein domains and the effect of swapping mutagenesis on the domain stabilities of multi and single domain proteins.
(a) Structure based sequence alignment between single domain-homologues and multi-domain protein domains were generated, and used to calculate the propensity of interface mutations (See methods). Observe the preferences for polar to apolar substitution frequencies in going from single-domain homologues to MD_doms. For e.g. the propensity of W to K substitutions in the interface. (b) The MD mutants represents the distribution of single point mutations (n = 5019), where each of the non-conserved interfacial residue was mutated to the corresponding residue found in the respective single domain homologue in the structure based sequence alignments. The 1D_hom mutants represents a set of single point mutations (n = 5019) on single domain proteins where the residues corresponding to the interface in the multi-domain protein homologue were mutated to those found in the MD_doms in the structure based sequence alignments. MD_mutants were significantly less destabilizing (Paired t-test; t = 12.60; df = 5018; P < 0.001) and show no relation (Spearman rank correlation test: r2 = −0.067; P = 1.50E-06) to stability effects in 1D_hom mutants.