Abstract
Transport of mRNAs to diverse neuronal locations via RNA granules serves an important function in regulating protein synthesis within restricted sub-cellular domains. We recently detected the Huntington's disease protein huntingtin (Htt) in dendritic RNA granules; however, the functional significance of this localization is not known. Here we report that Htt and the huntingtin-associated protein 1 (HAP1) are co-localized with the microtubule motor proteins, the KIF5A kinesin and dynein, during dendritic transport of β-actin mRNA. Live cell imaging demonstrated that β-actin mRNA is associated with Htt, HAP1, and dynein intermediate chain in cultured neurons. Reduction in the levels of Htt, HAP1, KIF5A, and dynein heavy chain by lentiviral-based shRNAs resulted in a reduction in the transport of β-actin mRNA. These findings support a role for Htt in participating in the mRNA transport machinery that also contains HAP1, KIF5A, and dynein.
Huntington's disease (HD) is a progressive neurodegenerative disorder characterized by the death of striatal neurons in the brain. The mutation that causes HD is an expansion of the polymorphic CAG repeats encoding polyglutamines in the huntingtin (Htt) protein1. Although the normal functions of Htt remain controversial, Htt has been shown to promote cargo transport along microtubules in axons2,3. Altered axonal transport by mutant Htt of cargos such as BDNF, critical for the survival of neurons, has been proposed to contribute to the pathogenesis of HD3. In Drosophila, reduction of endogenous Htt or expression of polyQ-expanded Htt exon 1 resulted in disruption of axonal transport implicating its role in this pathway4.
In addition to transporting proteins, a large number of mRNAs synthesized in the soma are transported as part of ribonucleoprotein particles (RNPs) to distal sites in neurons. Studies suggest that targeting of translationally silenced mRNAs to diverse intracellular locations where translation machinery resides allows regulated synthesis of select proteins in spatially restricted domains5,6,7. The local translation of mRNA in response to synaptic activity has been proposed to play a major role in synaptic plasticity and contribute to long-lasting changes in synaptic strength8,9. Thus, determination of the mechanism of mRNA transport and local translation is critical to understanding synaptic regulation.
Proteins involved in neuronal mRNA transport have been extensively studied (reviewed in10,11). They include heterogeneous nuclear ribonucleoprotein (hnRNP) A2, staufen, fragile X mental retardation protein (FMRP), and zipcode-binding protein 1 (ZBP1), among others. The mRNAs transported appear to contain targeting sequences in the 3′UTR. The examples include mRNA for β-actin, microtubule-associated protein 2 (MAP2), Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα), and Arc12,13,14,15,16. Neuronal mRNAs associate with a set of proteins to form RNPs, which are heterogeneous and dynamic in composition. Several types of RNPs have been reported: transport RNPs, stress granules (SGs), processing bodies (P-bodies), and dendritic P-body-like structures17. Large RNA transport granules contain dendritically localized mRNAs, ribosomes, RNA binding proteins such as staufen, motor proteins such as kinesin, and translation factors. SGs serve as sites for temporary arrest of mRNA translation in response to stress signals, while P-bodies function to store and/or degrade mRNAs. P-bodies also contain microRNAs and the RISC, which mediates post-transcriptional gene silencing. The relationship between different RNA granules remains unclear. Several proteins appear to be present in different types of RNA granules, suggesting heterogeneity/overlap among different RNPs; however, a recent study has shown distinct mRNAs transported by distinct RNA granules, implying a mechanism that confers specificity18,19. Another study found no overlap between transport RNPs and P-bodies although docking of these RNPs were observed20. It is possible that P-bodies function to maintain mRNAs in a repressed state until the synapses are activated, triggering transport and local translation of specific mRNAs. Finally, ATP-dependent motor proteins (kinesin and dynein) are involved in transport of mRNA along microtubules21,22,23.
We recently reported that Htt associates with Argonaute 2 (Ago2) at cytoplasmic P-bodies in non-neuronal cells and contributes to small RNA-mediated gene silencing24. We subsequently demonstrated Htt playing a role in mRNA trafficking in neurons25. In the current study we have characterized Htt and its relationship with neuronal mRNA transport machinery and local protein synthesizing machinery in dendrites. The newly defined function of Htt could lead to the uncovering of new pathogenic mechanisms of mutant Htt and potentially new therapeutic targets for HD.
Results
Htt is involved in β-actin mRNA transport in cultured neurons and brain sections
A combined approach of high-resolution FISH and immunofluorescent protein staining was used to study the distribution of β-actin mRNA and its association with Htt in cultured cortical neurons. β-actin mRNA-containing granules (ribonucleoprotein particles or RNPs) were observed in all compartments of cortical neurons (DIV9) including the soma, dendrites, and axons (Fig. 1a). To determine the fraction of β-actin RNPs potentially engaged in protein synthesis in the dendrite, neurons were probed for β-actin mRNA together with a ribosome marker ribosomal protein 6 (RPS6, Fig. 1b). 9.8% of β-actin RNPs co-localized with RPS6 (n = 235), suggesting that about 10% of β-actin mRNA is subject to local translation at steady state in cultured neurons.
Figure 1. Htt is involved in the dendritic transport of β-actin mRNA in rat cortical neurons.
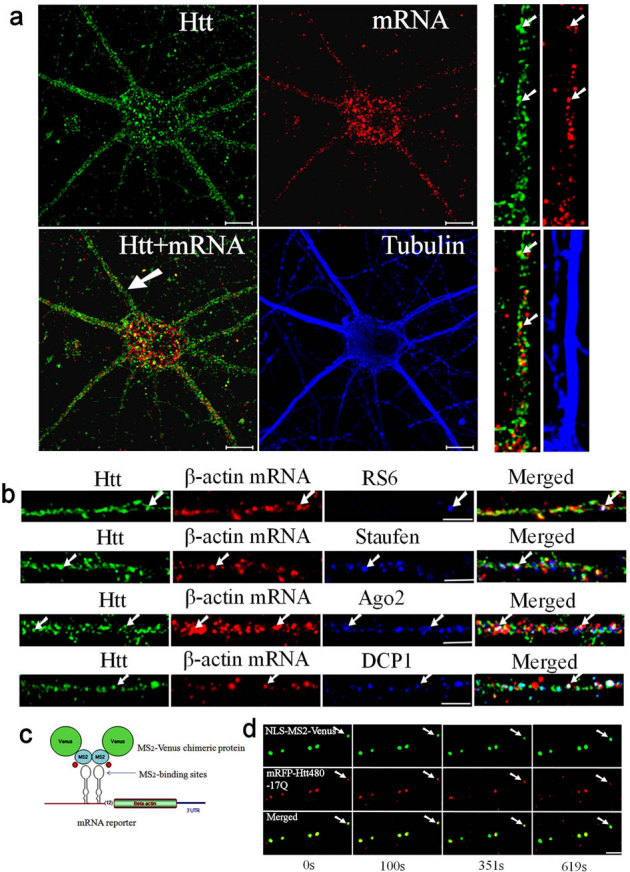
(a) Co-localization of Htt and β-actin mRNA in cortical neurons (DIV 9). Htt, β-actin mRNA, and α-tubulin are shown in green, red, and blue, respectively. The panels at right are high-magnification views of a single dendrite pointed with a white arrow in the merged image. The arrows indicate the co-localization of Htt and mRNA. Scale bar: 10.0 μm. (b) Htt, β-actin mRNA, and indicated RNP-associated protein (RPS6, staufen, Ago2, DCP1) are shown in green, red, and blue, respectively. Co-localization of Htt/mRNA, Htt/RNP-associated protein, and mRNA/RNP-associated protein is shown in yellow, cyan, and magenta, respectively, in the merged panel. The arrows indicate the co-localization of Htt, mRNA, and RNP-associated protein (appear as white). The left part of each image is the proximal part of the dendrite. Scale bar: 5.0 μm. (c) Schematic representation of the MS2-Venus reporter system for the visualization of β-actin mRNA in live cortical neurons. (d) Co-trafficking of Htt with β-actin mRNA in rat cortical neurons. β-actin mRNA is shown in green and co-transfected mRFP-Htt480-17Q in red. Four cropped images from a time-lapse series captured over 652.5 seconds are shown. The left part of each image is the proximal part of the dendrite. The arrows indicate one retrogradely moving RNA granule with Htt in the neuron. The distance that the granule traveled was 2.78 μm. Scale bar: 5.0 μm.
Htt was previously found to co-localize with mRNAs/RNPs in dendrites25. We immunostained neurons for Htt after FISH, and observed Htt co-localize with β-actin RNPs in all compartments (Fig. 1a). In the dendrites of DIV 9 neurons, 40% of β-actin RNP co-localized with Htt (n = 200). Studies have shown the presence of different types of RNA processing and storage centers in neurons, many of which are likely involved in mRNA transport10. Antibodies to known markers of neuronal RNPs were used to determine the relationship between Htt and neuronal RNPs. In addition to the ribosomal marker RPS6, Htt co-localized with β-actin RNP and the RNA-binding protein staufen (Fig. 1b). 21.4% of β-actin RNP co-localized with staufen (n = 206) and 56.8% of the β-actin-staufen RNP co-localized with Htt. We also found Htt co-localize with RNPs containing β-actin mRNA and Ago2 (Fig. 1b). 42.1% of β-actin RNP co-localized with Ago2 (n = 202) and 55.3% of the β-actin-Ago2 RNP co-localized with Htt. We next examined if Htt was present in RNPs containing a processing (P)-body marker DCP1 and β-actin mRNA (Fig. 1b). 33.3% of β-actin RNP co-localized with DCP1 (n = 207); 65.2% of the β-actin-DCP1 RNP co-localized with Htt. Co-localization of β-actin mRNA with staufen, Ago2, and DCP1 was also observed in the brain sections from the rat cortex and hippocampus (see Supplementary Fig. S1 online). Therefore, we have found Htt associated with RNPs containing β-actin mRNA and four distinct proteins in cultured neurons and brain sections. The four proteins, RPS6, staufen, Ago2, and DCP1, represent components of different RNA granules. Although we do not know the extent of overlap between different RNPs, Htt appears to be a part of multiple RNPs involved in mRNA transport.
The β-actin mRNA has a well-defined 3′UTR sequence that contains a dendritic targeting signal termed zipcode26. The dynamics of β-actin mRNA trafficking was next investigated in live neurons using a fluorescent protein reporter system. The MS2 system developed by the Kosik group27 is widely used to visualize the movement of mRNA in neurons. The β-actin mRNA reporter plasmid was constructed by inserting upstream of the β-actin cDNA with the zipcode sequence, multiple binding sites for the bacteriophage MS2 protein (Fig. 1c). Co-transfection of a plasmid expressing NLS-MS2-Venus with the reporter into DIV 5 primary rat cortical neurons permitted visualization of β-actin mRNA by live-cell imaging. A representative result demonstrating the trafficking dynamics of β-actin RNA is summarized in Supplementary Table S1 online. The average velocity of mRNA granules measured was 0.0294 ± 0.0215 μm/s, which is comparable to our previously recorded values for trafficking mRNAs visualized by SYTO RNASelect (average velocity: 0.04 μm/s)25.
Studies have shown that Htt contributes to the intracellular trafficking of cargos in axons3; however, there is no report of Htt participating in mRNA trafficking in neurons. To demonstrate co-trafficking of Htt with mRNA, we transfected neurons with three plasmids: MS2-β-actin mRNA reporter, NLS-MS2-Venus, and mRFP-Htt480-17Q. Live cell imaging found Htt co-traffic with β-actin mRNA (as visualized by NLS-MS2-Venus) in both directions (anterograde and retrograde). Representative results are shown in Fig. 1d, and Supplementary Video 1 online. A negative control in which mRNA reporter was omitted showed the Venus signal mostly restricted to the nucleus presumably due to the presence of the NLS (see Supplementary Fig. S2 online). This result suggests that dendritic co-localization of NLS-MS2-Venus with mRFP-Htt480-17Q is dependent on MS2-β-actin mRNA.
To provide additional evidence that Htt plays a role in β-actin mRNA transport, we depleted endogenous Htt with shRNA and examined β-actin mRNA localization by microscopy. We have previously demonstrated successful knockdown of Htt with the same shRNAs25. Cortical neurons were infected with a lentivirus expressing scrambled shRNA (control), shRNA-Htt-1, or shRNA-Htt-2, and probed at DIV 8 for endogenous Htt protein and β-actin mRNA. Upon knockdown of Htt, concomitant decrease in β-actin mRNA levels was observed (Fig. 2a, b). In addition, the percentage of β-actin mRNA in the soma (soma β-actin mRNA/total β-actin mRNA, i.e., intensity of entire neuron) increased significantly after the knockdown of Htt (Fig. 2c, d), supporting the idea that Htt is involved in the dendritic transport of β-actin mRNA. Although we cannot eliminate the possibility that changes in β-actin mRNA is due to dysfunctional neurons caused by depletion of Htt, neurons knocked down for Htt looked similar to control neurons after 96 hrs, indicating that Htt knockdown does not have detrimental effects on neurons during the course of the experiment (see Supplementary Fig. S3 online).
Figure 2. Knockdown of Htt decreases β-actin mRNA levels in rat neurons leading to accumulation in the soma.
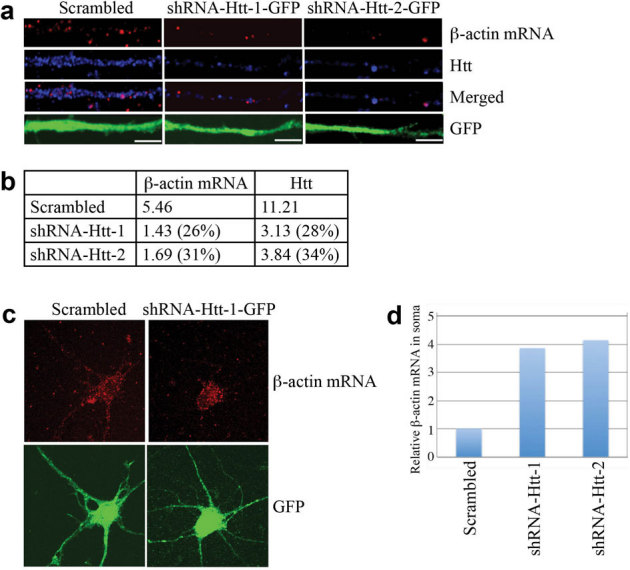
(a) DIV 4 cortical neurons were infected with lentivirus expressing scrambled shRNA, shRNA-Htt-1, or shRNA-Htt-2, and probed at DIV 8 for β-actin mRNA (red), Htt (blue), GFP (green, with anti-GFP antibody). Representative images from each experiment are shown. The left part of each image is the proximal part of the dendrite. Scale bar: 5.0 μm. (b) Relative intensity of Htt and β-actin mRNA shown in (a), normalized to the area of GFP staining, which represents size of dendrite. The percent shown in parentheses indicates extent of knockdown for Htt, and reduction in signal for β-actin mRNA relative to scrambled. (c) Representative images of β-actin mRNA and GFP in a whole neuron after knockdown of Htt. Scale bar: 10.0 μm. (d) Quantitative analysis of enrichment of β-actin mRNA in the soma shown in (c). Intensity of β-actin mRNA in the soma was divided by total β-actin mRNA. Control sample (infected with lentivirus carrying scrambled shRNA) was set to 1. n = 2.
HAP1 is required for dendritic targeting of β-actin mRNA
Htt interacts with HAP1, which plays an important role in intracellular cargo transport28. We next examined the role of HAP1 in the transport of β-actin mRNA. DIV 9 neurons were probed for β-actin mRNA together with HAP1 and Htt (Fig. 3a). We found 45.4% of β-actin mRNA to co-localize with HAP1 (n = 196); 60.6% of β-actin RNP that co-localized with HAP1 also co-localized with Htt. Co-localization of β-actin RNPs with Htt and HAP1 was also observed in the brain sections from the rat hippocampus (see Supplementary Fig. S4 online). Therefore, HAP1 is also present in β-actin RNPs and may be involved in mRNA transport in neurons. To investigate this, cortical neurons were co-transfected with MS2-β-actin mRNA reporter plasmid, NLS-MS2-Venus, and DsRed2-HAP1, a fusion of HAP1 with a red fluorescent protein. Like Htt (Fig. 1d), HAP1 was found to co-traffic with β-actin mRNA, both anterogradely and retrogradely. Representative results are shown in Fig. 3b, 3c, and Supplementary Video 2 online. Knockdown of endogenous HAP1 further supported these findings. DIV 4 cortical neurons were infected with a lentivirus expressing scrambled shRNA or shRNA-HAP1, and probed at DIV 8 for HAP1 and β-actin mRNA (Fig. 3d, e). Depletion of HAP1 resulted in substantial reduction of dendritic β-actin mRNA, indicating its role in mRNA transport. Like Htt, knockdown of HAP1 did not have detrimental effects on neurons during the course of the experiment (data not shown).
Figure 3. HAP1 is involved in the dendritic targeting of β-actin mRNA.
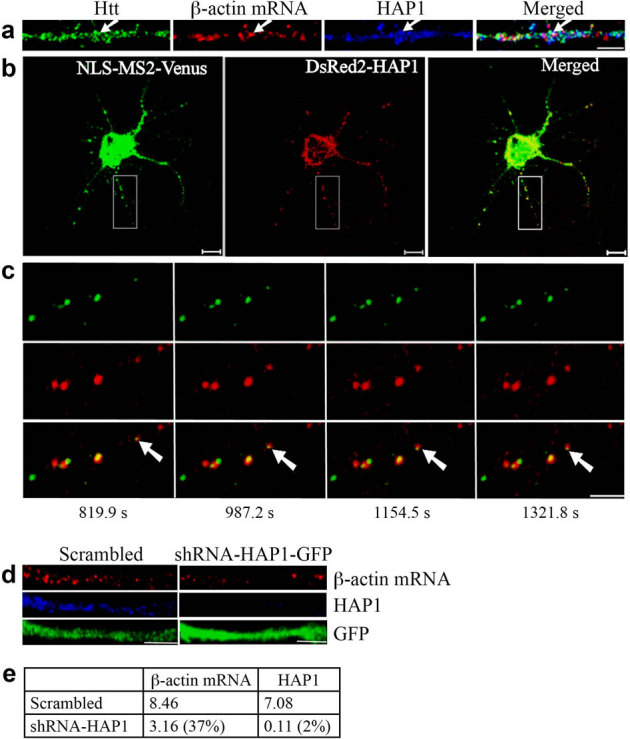
(a) Co-localization of Htt, β-actin mRNA, and HAP1 in rat cortical neurons (DIV 9). Htt, β-actin mRNA, and HAP1 are shown in green, red, and blue, respectively. The left part of each image is the proximal part of the dendrite. The arrows indicate the co-localization of Htt, β-actin mRNA, and HAP1. Scale bar: 5.0 μm. (b) Co-trafficking of HAP1 with β-actin mRNA in rat cortical neurons (DIV 6). β-actin mRNA is seen in green and HAP1 in red. HAP1 co-localizing with β-actin mRNA is in yellow in the merged panel. Scale bar: 10.0 μm. Part of the boxed area is shown at a high-magnification in (c). (c) Four cropped images from a time-lapse series captured over 1656.5 seconds. The left part of each image is the proximal part of the dendrite. The arrows indicate retrograde movement of one mRNA granule with HAP1 in the neuron. The distance that the granule traveled was 4.03 μm. Scale bar: 5.0 μm. (d) Knockdown of HAP1 decreases β-actin mRNA levels in rat neurons. DIV 4 cortical neurons were infected with lentivirus expressing scrambled shRNA or shRNA-HAP1 and probed at DIV 8 for β-actin mRNA (red), HAP1 (blue), and GFP (green). The left part of each image is the proximal part of the dendrite. Scale bar: 5.0 μm. (e) Quantitative analysis of the intensity of β-actin mRNA and HAP1 normalized to GFP. The percent shown in parentheses indicates extent of knockdown for HAP1, and reduction in signal for β-actin mRNA relative to scrambled.
β-actin mRNA transport is microtubule-dependent
Dendritic mRNA transport has been shown to involve microtubules. Thus, we examined the role of microtubules in Htt-mediated β-actin mRNA transport. FISH and immunostaining were performed to assess the potential association of Htt/β-actin RNP with microtubules stained with anti-βIII-tubulin antibody (Fig. 4a, top). Most of the β-actin mRNA was found associated with microtubules in the dendrite. Disruption of microtubules following a 30-minute treatment with 2 μg/ml nocodazole had little effect on the distribution of dendritic β-actin mRNA during the short time course (Fig. 4a, bottom). However, the dynamics of β-actin mRNA movement changed significantly after the nocodazole treatment (Fig. 4b). In the absence of the drug, bi-directional movement of a subset of β-actin RNPs was observed (see Supplementary Video 3 online); however, treatment of neurons with nocodazole resulted in RNPs that appeared stationary or subject to Brownian motion (see Supplementary Video 4 online). These results suggest that the transport of Htt/β-actin RNP is dependent on microtubules.
Figure 4. Transport of mRNA in dendrites is microtubule-dependent.
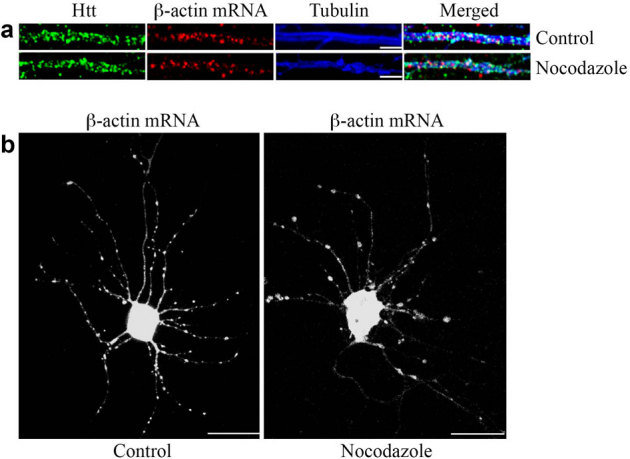
(a) The distribution of β-actin mRNA in the dendrites of DIV 9 control neuron and neuron treated with nocodazole. In the control neuron, β-actin mRNA is associated with microtubules. The staining of mRNAs does not change significantly after a 30-minute treatment with 2 μg/ml nocodazole. Scale bar: 5.0 μm. (b) Visualization of β-actin mRNA in the control neuron and neuron after nocodazole treatment (2 μg/ml, 30 minutes). Scale bar: 10.0 μm.
Kinesin-1 and dynein are involved in β-actin mRNA transport
The HAP1-KIF5 complex was recently shown to be involved in the delivery of GABAA receptors to inhibitory synapses, and mutant Htt impaired synaptic inhibition by disrupting receptor transport29. To investigate whether motor proteins kinesin-1/KIF5A and dynein are involved in Htt-mediated dendritic targeting of β-actin mRNA, FISH and immunostaining methods were used to determine the association of the motor proteins with β-actin mRNA and Htt. First, KIF5A was found to co-localize with β-actin mRNA and Htt in dendrites (Fig. 5a). 30.1% of β-actin mRNA co-localized with KIF5A (n = 136); 19.1% of β-actin mRNA co-localized with Htt and KIF5A (n = 136). Second, dynein heavy chain (DHC) was found to co-localize with β-actin mRNA and Htt in dendrites (Fig. 5a). 31.0% of β-actin mRNA co-localized with dynein (n = 229); 13.1% of β-actin mRNA co-localized with Htt and dynein (n = 229). Third, β-actin mRNA was found to co-localize with KIF5C and dynein HC in dendrites (Fig. 5a). 31.4% of β-actin mRNA co-localized with KIF5C (n = 210), whereas 15.3% of β-actin mRNA co-localized with kinesin-1 and dynein (n = 183), similar to the co-localization percentage of β-actin mRNA with KIF5A (19.1%, see above). Similar co-localization patterns were observed in brain sections from rat cortex and hippocampus (see Supplementary Fig. S5 online). Fourth, the co-localization of p150Glued, dynein, and β-actin mRNA was observed (Fig. 5a). Finally, we examined the distribution of Htt phosphorylated at S421, a modification reported to function as a molecular switch in the regulation of anterograde and retrograde trafficking in the axon3. DIV 9 cortical neurons were probed for β-actin mRNA, Phos-Htt, and total Htt (Fig. 5b). We found that 20.3% of β-actin mRNA co-localized with Phos-Htt (n = 162). Because 40.0% of β-actin mRNA co-localized with total Htt, we surmised that about one-half of co-localizing Htt was phosphorylated at S421 (Fig. 5b).
Figure 5. Motor proteins kinesin-1 and dynein are involved in the dendritic transport of β-actin mRNA.
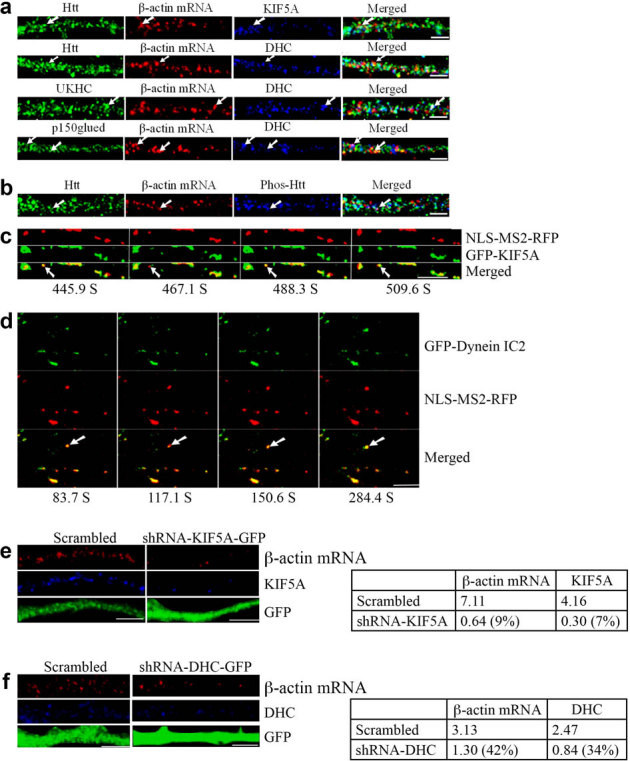
The left part of each image is the proximal part of the dendrite. DHC: dynein heavy chain. Scale bar: 5.0 μm. (a) KIF5A and dynein co-localize with β-actin mRNA in rat cortical neurons (DIV 9). The arrows indicate the co-localization of three targets. UKHC: kinesin-1 heavy chain. (b) Co-localization of Htt, β-actin mRNA, and Phos-Htt in rat cortical neurons (DIV 9). (c) Live cell co-trafficking of KIF5A with β-actin mRNA in rat cortical neurons was imaged over 509.6 seconds. β-actin mRNA (detected by NLS-MS2-RFP) is in red and KIF5A (GFP) in green. The arrow indicates an mRNA granule that moves anterogradely along the dendrite. The distance that the granule traveled was 1.39 μm. The velocity of the granule is 0.013 μm/s. (d) Co-trafficking of DIC with β-actin mRNA in rat cortical neurons was imaged live over 1040.4 seconds. β-actin mRNA (detected by NLS-MS2-RFP) is in red and DIC (EGFP) in green. The arrow indicates an mRNA granule that moves retrogradely along dendrites. The distance that the granule traveled was 1.57 μm. DIC: dynein intermediate chain. (e–f) Knockdown of KIF5A or DHC decreases β-actin mRNA levels in rat neurons. DIV 4 cortical neurons were infected with a lentivirus expressing scrambled shRNA, shRNA-KIF5A, or shRNA-DHC and probed at DIV 8 for β-actin mRNA (red), KIF5A or DHC (blue), and GFP (green). The left part of each image is the proximal part of the dendrite. The right panel shows quantitative analysis of the normalized intensity. The percent shown in parentheses indicates extent of knockdown for KIF5A or DHC, and reduction in signal for β-actin mRNA relative to scrambled.
To demonstrate a direct involvement of KIF5A in β-actin mRNA transport, cortical neurons were co-transfected with plasmids encoding MS2-β-actin mRNA reporter, NLS-MS2-RFP, and GFP-KIF5A. KIF5A was found to co-traffic anterogradely with β-actin mRNA. One representative result is shown in Fig. 5c, and Supplementary Video 5 online. To investigate the role of dynein (intermediate chain, DIC2) in β-actin mRNA transport, cortical neurons were co-transfected with plasmids encoding MS2-β-actin mRNA reporter, NLS-MS2-RFP, and GFP-DIC. DIC was found to co-traffic retrogradely with β-actin mRNA. One representative result is shown in Fig. 5d, and Supplementary Video 6 online.
We also examined the effect of knockdown of kinesin-1 and dynein on the dendritic targeting of β-actin mRNA. DIV 4 cortical neurons were infected with a lentivirus expressing scrambled shRNA, shRNA-KIF5A, or shRNA-DHC and probed at DIV 8 for KIF5A or DHC, and β-actin mRNA (Fig. 5e, f). The depletion of KIF5A or DHC resulted in substantial reduction of β-actin mRNA in the dendrite, consistent with the above findings that KIF5A and DHC are involved in the transport of β-actin mRNA.
Interaction of Htt and other proteins in mRNA transport
Htt is thought to form a network of interactions with motor proteins involved in intracellular vesicular trafficking2. To investigate the presence of a similar network in the transport of mRNA, we analyzed the effect of knockdown of one protein on the levels of other proteins in the transport machinery. Several interesting findings were observed (Table 1). First, knockdown of Htt had no effect on the levels of HAP1, KIF5A, and dynein in the dendrite. Second, knockdown of HAP1 had no effect on the levels of Htt, but decreased the levels of KIF5A and dynein in the dendrite. Third, knockdown of KIF5A decreased the levels of all three proteins – Htt, HAP1, and dynein in the dendrite. Fourth, knockdown of DHC had no effect on the Htt levels. However, it resulted in decreased levels of HAP1 and increased levels of KIF5A. We think these proteins are co-regulated because they associate with one another to form large protein transport machinery. Depleting one component by knockdown could destabilize the complex leading to loss of other components.
Table 1. Effect of knockdown on the expression of proteins in the transport machinery.
| Htt | HAP1 | KIF5A | Dynein | |
|---|---|---|---|---|
| shRNA-Htt-1 | ------ | → | → | → |
| shRNA-HAP1 | → | ------ | ↓(16.4%) 0.84/5.13 | ↓(32.3%) 0.94/2.91 |
| shRNA-KIF5A | ↓(31.1%) 1.96/6.31 | ↓(35.2%) 1.60/4.54 | ------ | ↓(17.5%) 0.51/2.91 |
| shRNA-DHC | → | ↓ (33.3%) 1.51/4.54 | ↑ (160%) 8.20/5.13 | ------ |
→ No significant changes.
The percentage was calculated as follows: normalized intensity (underlined index number) after knockdown of target protein with indicated shRNA, was divided by normalized intensity (italicized index number) of the same target with scrambled shRNA. The area of anti-GFP staining in 30 μm dendrites was used for normalization.
Studies found neuronal activity to induce local translation of dendritic mRNAs to proteins; thus, we investigated whether mRNAs of the above proteins are present in dendrites (see Supplementary Fig. S6 online). The mRNA of Htt, HAP1, ZBP1, KIF5A and DIC were all found in dendrites and co-localized with endogenous Htt, suggesting that Htt is involved in transport of its own mRNA as well as mRNAs encoding components of transport machineries.
Zipcode-ZBP1 pathway of β-actin mRNA transport
The dendritic targeting of β-actin mRNA has been demonstrated to require a 54-nt sequence in the 3′UTR15, which binds to an RNA binding protein ZBP130. Our study so far has revealed that β-actin mRNA transport involves Htt and proteins associated with different RNA granules as well as microtubules. Thus, we investigated the localization of ZBP1, a known β-actin mRNA binding protein, to determine its relationship with Htt. 31.4% of β-actin mRNA co-localized with ZBP1 (n = 140) (Fig. 6a) while 30.1% of ZBP1 co-localized with β-actin mRNA (n = 146). Significantly, 79.5% of the β-actin mRNA-ZBP1 complex (25.0% of total β-actin RNP) co-localized with Htt indicating that Htt is involved in the ZBP1-mediated β-actin mRNA transport.
Figure 6. The zipcode sequence in the 3′UTR of β-actin mRNA is sufficient for dendritic targeting and co-localization with Htt in rat neurons.
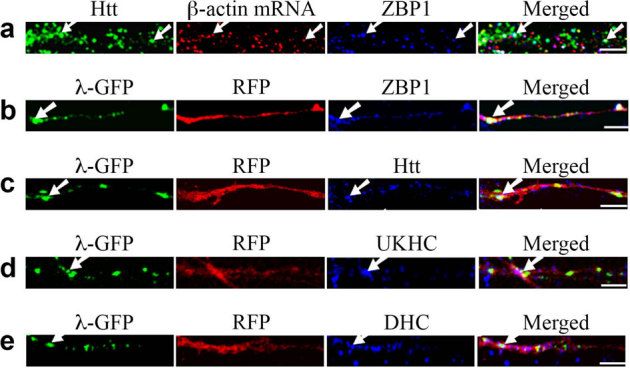
The left part of each image is the proximal part of the dendrite. Scale bar: 5.0 μm. (a) cortical neurons (DIV 9) were probed for Htt (green), β-actin mRNA (red), and ZBP1 (blue). The arrows indicate the co-localization of Htt, β-actin mRNA, and ZBP1. (b–e) β-actin-zipcode mRNA is visualized by co-transfection of RFP-4xboxB-β-actin-zipcode reporter and λN-GFP, which binds to the 4xboxB sequence. RFP and mRNA are shown in red and green, respectively. Co-localization of mRNA (green) and endogenous proteins (blue) of the transport machinery is indicated by arrows. Co-localization of the translated RFP and endogenous ZBP1, Htt, kinesin-1 (UKHC), and dynein HC are shown in (b), (c), (d), and (e), respectively. Although λN-GFP also represents RFP mRNA, translated RFP diffuses within the dendrite resulting in the loss of punctate pattern of mRNA. DHC: dynein heavy chain.
To confirm that the zipcode sequence is sufficient for the dendritic targeting of β-actin mRNA and co-localization with Htt, a reporter system similar to MS2, but based on the bacteriophage lambda boxB sequence and the binding protein λN fused to GFP, was used31. DIV 4 neurons were transfected with plasmids encoding RFP-4xboxB-β-actin zipcode mRNA reporter and λN-GFP, and probed for Htt and other proteins 24 hours after transfection. The β-actin zipcode-containing mRNA visualized by λN-GFP co-localized with endogenous ZBP1, Htt, kinesin-1, and dynein (Fig. 6b-e) indicating that the zipcode sequence of β-actin was sufficient for Htt-mediated transport. We also performed an experiment in which mRFP-Htt480-17Q and ZBP1-GFP were co-transfected and the movement of co-localizing RNP measured. Htt was found to co-traffic anterogradely and retrogradely with ZBP1 (data not shown).
Co-fractionation of Htt with motor proteins
To obtain biochemical evidence supporting the co-localization studies, we prepared mouse brain homogenates and separated proteins from the lysate through 10-40% glycerol gradient (Fig. 7a). We found Htt co-fractionates with microtubule motors, DHC, dynactin 1, KIF5A, HAP1, as well as with Rps6. The results are consistent with the molecular associations demonstrated in the co-localization experiments.
Figure 7. Htt co-fractionates with microtubule motors and Rps6 in mouse brain.
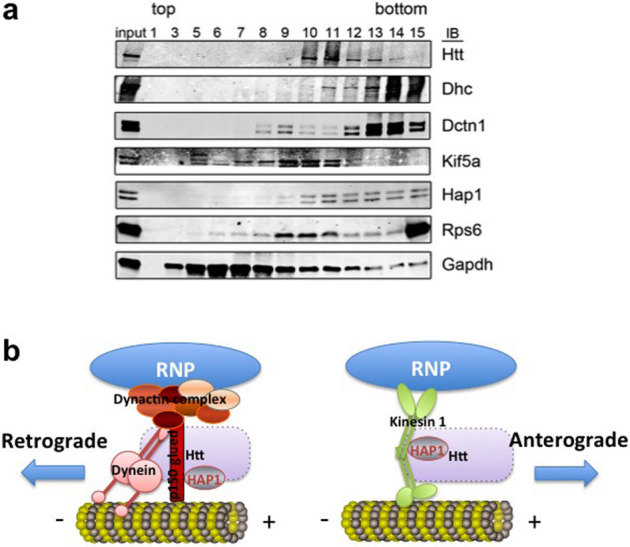
(a) Western blot of glycerol gradient fractions for indicated proteins (at right). Increasing numbers correspond to lower (denser) position in the gradient. The material loaded onto the gradient is represented in the input lane (3%). Dhc, dynein heavy chain. Dctn 1, dynactin 1. (b) Proposed model of Htt-mediated RNP transport in the dendrite. Htt/HAP1/dynein/dynactin complex is involved in retrograde transport of RNP, whereas Htt/HAP1/kinesin is involved in anterograde transport of RNP. Exact placement of Htt is not known (indicated by a dashed rectangle).
Discussion
Based on our findings, we propose a model (Fig. 7b) for dendritic β-actin mRNA transport machinery that include: (a) Htt and HAP1, (b) specific mRNA-binding proteins, such as ZBP1, and general mRNA-binding proteins/mRNA-associated proteins, such as staufen, Ago2, and DCP1, (c) motor proteins kinesin-1 and cytoplasmic dynein, (d) microtubules. Although the model depicts these components to be present within a complex, the transport machinery is likely heterogeneous and dynamic in composition as suggested by many reports in the literature.
In this study, trafficking patterns of β-actin RNPs were analyzed by live-cell imaging. The β-actin RNP travels at an average velocity of 0.0294 μm/s. This value is comparable with our previously recorded values of mRNA trafficking visualized by SYTO RNASelect, which detects all mRNAs and thus likely represents the trafficking pattern of endogenous RNPs25. This value is also comparable with the reported velocity of slow axonal transport in neurons (0.004 μm/s – 0.04 μm/s,32), although there is a recent paper reporting much faster transport rates of RNPs (∼0.4 μm/s) by microinjection of fluorescently labeled mRNA into live neurons19.
Htt participates in cargo transport, and pathogenic polyQ-containing Htt perturbs axonal and dendritic cargo transport3,33. Htt associates with various proteins that may play a role in intracellular trafficking34. In particular, Htt interacts with HAP1, a protein that is transported in axons and associates with the p150 glued dynactin subunit, an essential component of the dynein/dynactin microtubule-based motor complex3. However, a role for Htt in mRNA trafficking has not been reported. Using high-resolution co-localization analysis and live-neuron imaging, we demonstrated in cultured neurons and rat brain sections that Htt and HAP1 are involved in mRNA targeting and trafficking. In addition, knockdown of Htt in cultured primary neurons decreased the levels of β-actin mRNA (Fig. 2) and total mRNA25. The percentage of β-actin mRNA in the soma increased after Htt knockdown, suggesting mRNA that is no longer transported to the dendrite and other distal parts of the neurons accumulate in the soma. Therefore, we report for the first time that wild-type Htt is involved in β-actin mRNA transport in neurons, a process implicated in modulating dendrite development and synaptic plasticity35. The precise mechanism by which Htt and HAP1 contribute to mRNA transport remains to be determined.
Kinesin-1 and dynein are two important components of mRNA transport machinery in dendrites21,23. Hirokawa and colleagues isolated a large detergent-resistant RNase-sensitive granule (∼1000S in size) from mouse brain that bound to conventional kinesin KIF5, and showed that transport of RNA-containing granules in dendrites required the C-terminal tail of KIF522, although the binding partners of KIF5 remain unknown. Interestingly, it was reported that dynein light chain binds to a 3′UTR sequence of parathyroid hormone mRNA implicating microtubules in RNA transport36. Our study suggests that Htt-HAP1 mediates dendritic transport of β-actin mRNA as an RNP complex associated with the motor proteins kinesin and dynein.
The dendritic transport of RNPs is a highly regulated process with RNPs moving anterogradely or retrogradely, depending on the molecular motors, kinesin and dynein, respectively, which propel RNPs along microtubules. In our study, 31.4% of β-actin RNPs co-localized with kinesin-1, and 31.0% of β-actin RNPs co-localized with dynein, whereas 15.3% of β-actin RNPs co-localized with kinesin-1 and dynein. These data indicate that not all RNPs are associated with either one or both motor proteins, which differ from the report that all RNPs are associated with both kinesin-1 and dynein21.
In vitro studies have demonstrated that dynein and kinesin can function independently, and that the direction of transport of a given cargo along microtubules depends on the nature of the molecular motor present37. By contrast, in vivo, high levels of coordination are necessary to ensure proper transport of a given cargo to the right place. In our study, KIF5A and DIC seemed to co-traffic with β-actin RNP mostly anterogradely and retrogradely, respectively. However, some DIC-β-actin RNP complexes were found to undertake bi-directional movements in the dendrites indicating that the β-actin RNP might have associations with both motor proteins at the same time, and that certain “switch” mechanism might be responsible for the transition of the two directions of transport.
The phosphorylation of Htt at S421 has been reported to act as a molecular switch between anterograde and retrograde transport38. When Htt is phosphorylated, kinesin-1 is recruited to cargos and microtubules, thereby facilitating anterograde transport. By contrast, retrograde transport is favored in the absence of Htt phosphorylation at S421. In our proposed model, Htt-HAP1 also participates in this process and has an important regulatory function.
Although Htt interacts with the dynein intermediate chain, p150 glued, and kinesin light chain (KLC), mutant Htt does not affect the levels or the complex composition of Htt, HAP1, dynactin, and kinesin-1. Moreover, in contrast to the published data3, no changes in the biochemical behavior and potential interactions of Htt with HAP1 and motor protein complexes were observed in Hdh150Q/150Q brain extracts39. We have also performed similar studies on wild type and HD neurons and obtained similar results (data not shown). Only the dynein complex levels in the 25-μm distal segment of the HD neurons were slightly higher than those in wild type neurons. Furthermore, more RNPs are associated with KIF5A and dynein in HD neurons compared with wild type, indicating a higher affinity of RNPs with motor proteins (data not shown). If the mutant Htt impairs mRNA transport, then it may exert its effect by reducing the association of RNP-motor protein complexes with microtubules.
For most neuronally localized RNAs, the cis-acting sequences are not well defined, often being associated with RNA segments in the 3'-UTR. In the case of β-actin mRNA, the trafficking sequence is termed zipcode and the binding protein ZBP1. Approximately one-third of β-actin RNP and one-third of ZBP1 co-localized with β-actin RNP. This indicates that not all β-actin RNP is associated with ZBP1. ZBP1 can also bind other mRNAs with a similar zipcode sequence and mediate their transport in neurons. Our data show that a high percentage of the β-actin mRNA-ZBP1 complex (79.5%) co-localizes with Htt: this data (Fig. 6), and co-trafficking of zipcode sequence with Htt construct (Htt480-17Q), demonstrates a new role for Htt in β-actin mRNA transport.
Our previous study found Htt associated with Ago2 and P-bodies and contributed to RNA-mediated gene silencing24. It is thought that mRNAs are kept in repressed state during transport and become translationally competent upon synaptic activation. Thus, post-transcriptional processes such as transport and gene silencing are coupled, and dendritically targeted mRNAs may undergo a transition between the two states. The polyQ-expanded mutant Htt has been reported to alter axonal transport of cargo proteins such as BDNF; it remains to be seen if mutant Htt affects dendritic mRNA transport and contributes to the pathogenesis of HD. If so, genetic and pharmacological manipulations40 that restore such defects could be used as potential therapies for HD.
Methods
Antibodies
The following antibodies were purchased from commercial sources: mouse anti-Htt (MAB2166) and rabbit anti-staufen (Millipore), rabbit anti-ribosomal protein S6 (RPS6, #2217, Cell Signaling Technology), rabbit anti-decapping protein 1 (DCP1) and rabbit anti-βIII-tubulin (Sigma), rabbit anti-huntingtin-associated protein 1 (HAP1, Abnova), rabbit anti-dynein heavy chain (dynein HC, Santa Cruz Biotechnology), mouse anti-p150Glued (BD Biosciences). Rabbit anti-green fluorescent protein (GFP) and mouse anti-GFP were obtained from UC Davis/NIH NeuroMab Facility, mouse anti-tGFP was purchased from OriGene Technologies, Alexa 488-conjugated, goat anti-mouse IgG was purchased from BD Pharmingen. DyLight 549-conjugated, goat anti-chicken IgY (IgG) and goat anti-mouse IgG were from Jackson ImmunoResearch Laboratories.
The following antibodies were kind gifts of investigators: rabbit anti-Ago2 (24, Ramin Shiekhattar, The Wistar Institute), rabbit anti-KIF5A22, and mouse anti-KIF5C41 (ubiquitous kinesin heavy chain, UKHC; Nobutaka Hirokawa, University of Tokyo, Japan), rabbit anti-zipcode-binding protein1 (ZBP142; Gary Bassell, Emory University), anti-phosphorylated huntingtin (pS42143; Michael Hayden, University of British Columbia).
Plasmids
The following plasmids were kind gifts of investigators: monomeric red fluorescent protein (mRFP)-Htt-480-17Q (Florian Then, Massachusetts General Hospital)44, DsRed2-HAP145, GFP-KIF5A (full-length; Don Arnold, University of Southern California), EGFP-dynein intermediate chain 2 (DIC2, full-length;46), NLS-MS2-Venus47, NLS-MS2-RFP48, pmRFP-4xboxB-β-actin Zipcode and p4λN22-mEGFP3-M931, ZBP1-GFP (Gary Bassell, Emory University).
The MS2-β-actin mRNA reporter construct was generated by first subcloning rat β-actin cDNA including the zipcode sequence into a CMV-based mammalian expression plasmid pCGT49, followed by the insertion of 12xMS2-binding sequence upstream of the β-actin cDNA.
Preparation of cryosections
Female wild-type Wistar rats (3–4 weeks old, Harlan Laboratories, Indianapolis, IN) were sacrificed by CO2 narcosis. Brain tissue was quickly removed, embedded in Tissue-Tek® optimal cutting temperature (OCT) compound (Sakura Finetek, Torrance, CA), and snap-frozen in liquid nitrogen. Cryosections (10 μm thick) were prepared using Reichert-Jung 2800e Cryostat (Leica Microsystems) and mounted on 12-mm cover slips (pre-washed and pre-coated with poly-L-lysine). All rats were maintained under veterinary supervision at New York University School of Medicine Animal Facility in accordance with the guidelines established by the NIH for the care of laboratory animals, and all procedures approved by Institutional Animal Care and Use Committee.
Preparation of nick-translated probes
Digoxigenin (DIG)-labeled DNA probes were generated using a DIG-Nick Translation Mix (Roche Applied Science) according to the manufacturer’s instructions. One μg of plasmid DNA (described above) and ZBP1-EGFP35 was used to generate probes as described50. Plasmids without any insert were used as controls, which gave no FISH signals.
Fluorescence in situ hybridization (FISH) of mRNA
The experiments were carried out essentially as described50. The neurons / brain sections were fixed with 4% paraformaldehyde (PFA) for 18 minutes at room temperature, treated with 0.25% Triton X-100 in PBS for 10 minutes, and washed 3 times with PBS. After a 10-minute rinse in 1x sodium chloride/sodium citrate (SSC), the sections were incubated for 1 hour at 37°C in 100 μl hybridization buffer as described50. The neurons/sections were then hybridized for 6 hours at 37°C with 1 μl nick-translated probe diluted in 100 μl hybridization buffer. After hybridization, sections were washed with 40% formamide/1x SSC for 30 minutes at 37°C with gentle shaking, followed by three 15-minute washes with 1x SSC with gentle shaking on an orbital shaker.
Detection of DIG-labeled probes and immunofluorescence staining
For immunostaining, neurons were fixed with 4% PFA in PBS for 18 minutes at room temperature. All dilutions and washes (3×10 minutes) between stages were performed in PBS unless otherwise stated. Neurons/sections were washed for 20 minutes in PBS, blocked with antibody dilution buffer [3% goat serum (Sigma) and 1% IgG-free BSA] for 1 hour, and incubated for 1 hour with primary antibodies followed by a 45-minute incubation with secondary antibodies. After immunolabeling, samples were mounted on glass slides with fluorescent mounting medium (DAKO). DIG-labeled probes were detected by incubating sections with chicken anti-DIG antibody (Immunology Consultants Laboratory), followed by incubation with DyLight 549-conjugated goat anti-chicken IgY (IgG).
Confocal microscopy
Confocal imaging was performed using an LSM META510 confocal scanning laser system on an Axiovert 200 M microscope (Carl Zeiss) as described50 using the instrument settings detailed in Supplementary Table S2 online. Typically, 25-μm-long segments (proximal part) of dendrites were selected for analysis. Puncta-like staining in neurons was counted manually, and co-localization events identified by examining co-localization in two or three split color channels. Punctae with more than 30% overlap were recognized as co-localization events. The percentage of co-localization was calculated by dividing the co-localization events by the total number of events. “n” refers to the number of granules or events (protein or mRNA granules) analyzed.
Culture and transfection of primary neurons
Rat cortical neurons were isolated and cultured as previously described51. Neurons at days in vitro (DIV) 4–5 were transfected with 1 μg of total plasmid DNA and 1 μl Lipofectamine 2000 (Invitrogen) in OptiMEM (Invitrogen) per 24 well, as recommended by the manufacturer. For double transfections, 0.5 μg of each construct was used; for triple transfections, 0.33 μg of each construct was used. Before each transfection, 50% of the culture medium was removed and later used to replace the medium after 1-hour incubation with the transfection mixture.
Live-cell imaging
At 18–24 hours post transfection, neurons were transferred to a Lab-Tek 2-well chamber cover glass (Nalge Nunc International) for live-cell imaging. Images were acquired with a Plan-Neofluar 40x/1.30 oil-immersion objective lens. Lower laser power (3% for Argon laser and 10% for HeNe1) was used to avoid photo-bleaching and toxic effects. A Tempcontrol 37-2 Digital Microscope Incubator system (PECON, Ulm, Germany) was used to keep cells alive. The frame time was 15 seconds, frame intervals 5.0 seconds, and frame size 1024x1024 pixels.
Images in the LSM program were exported as a series of JPEG files and imported to ImageJ 1.42 (http://rsb.info.nih.gov/ij/). The image stacks were then further processed and saved as uncompressed AVI files. Video files were then compressed with Virtual Dub (Version 1.9.0.0, www.virtualdub.org) and saved as normal AVI files. Cinepak Codec by Radius was used for compression.
Lentivirus-based gene knockdown
Lentiviruses expressing Htt shRNA-1, Htt shRNA-2, or scrambled shRNA have been described25. Viral particles expressing GIPZ lentiviral shRNAmir for HAP1, KIF5A, and cytoplasmic dynein 1 heavy chain 1 (DYNC1H1) were purchased from Open Biosystems/ThermoFisher. Cortical neurons were infected at DIV 4 and cultured for an additional 4 days. The neurons were then probed for β-actin mRNA and proteins of interest. The intensity of β-actin mRNA and proteins were measured with ImageJ and normalized to the area of GFP staining.
Glycerol gradient fractionation of mouse brain lysate
One wild-type post-natal day 15 mouse brain was homogenized in 2 ml of buffer containing 10 mM HEPES, pH 7.6, 1.5 mM MgCl2, protease inhibitors, and 40 units RNAsin (Promega) in a dounce homogenizer with 6 strokes pestle A and 10 strokes pestle B. Brain homogenate was incubated for 10 minutes on ice and buffer salt concentrations were adjusted by adding 1/9th volume of 300 mM HEPES, pH 7.6, 1.4 M KCl, 30 mM MgCl2, 0.25% Triton X-100. Sequential centrifugations at 1400 g, 14,000 g, and 100,000 g pelleted nuclei, large cytoplasmic membranes, and insoluble particulates, respectively. 1 mg of soluble protein from the 100,000 g centrifugation was loaded onto 5 ml of a 10–40% glycerol gradient in an isotonic buffer and spun for 14 hours at 43,000 rpm in an SW-55 rotor at 4°C. 300 µl fractions were collected, TCA-precipitated, and resuspended in 2x Laemmli sample buffer. 25% of each fraction was analyzed by SDS-PAGE followed by western blotting.
Author Contributions
BM and NT conceived the study and designed the experiments, BM, JNS, MSY, and BPC performed the experiments and evaluated the data, BM, MVC, and NT wrote the paper. All authors have read and approved the final manuscript.
Supplementary Material
SUPPLEMENTARY INFORMATION
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Video 5
Supplementary Video 6
Acknowledgments
This work was supported by the Hereditary Disease Foundation (NT), grants from the CHDI Foundation, Inc. (NT, MVC), a grant from the NIH (NS061917 to NT), and a Shared Instrumentation Grant from the NIH (S10 RR017970). We thank Ramin Shiekhattar for the AGO2 antibody, Nobutaka Hirokawa for the anti-KIF5A and anti-KIF5C (UKHC) antibodies, Gary Bassell for the anti-ZBP1 antibody and the ZBP1 plasmid, Michael Hayden for the anti-Phos-Htt antibody, Hiroko Bannai for the NLS-MS2-Venus plasmid, Jan Ellenberg for the λN reporter system, Trina Schroer for the EGFP-DIC2, Don Arnold for the GFP-KIF5A, Thomas Meier for the MS2-RFP, and Wayne Rasband for the ImageJ program.
References
- Borrell-Pages M., Zala D., Humbert S. & Saudou F. Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci 63, 2642–2660 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J. P. & Holzbaur E. L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol 19, 147–155 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier L. R. et al.. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118, 127–138 (2004). [DOI] [PubMed] [Google Scholar]
- Gunawardena S. et al.. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron 40, 25–40 (2003). [DOI] [PubMed] [Google Scholar]
- Bramham C. R. & Wells D. G. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 8, 776–789 (2007). [DOI] [PubMed] [Google Scholar]
- Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci 26, 7139–7142 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. C. & Zukin R. S. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci 26, 7131–7134 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. & Schuman E. M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273, 1402–1406 (1996). [DOI] [PubMed] [Google Scholar]
- Huber K. M., Kayser M. S. & Bear M. F. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science 288, 1254–1257 (2000). [DOI] [PubMed] [Google Scholar]
- Loya C. M., Van Vactor D. & Fulga T. A. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev 24, 625–635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M. & Kiebler M. A. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. The EMBO journal 30, 3540–3552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg A. et al.. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci 19, 8818–8829 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell G. J. et al.. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci 18, 251–265 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg A. et al.. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci 13, 1881–1888 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang H. L. et al.. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron 31, 261–275 (2001). [DOI] [PubMed] [Google Scholar]
- Lyford G. L. et al.. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14, 433–445 (1995). [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M., Macchi P. & Dahm R. Perplexing bodies: The putative roles of P-bodies in neurons. RNA Biol 5, 244–248 (2008). [DOI] [PubMed] [Google Scholar]
- Mikl M., Vendra G. & Kiebler M. A. Independent localization of MAP2, CaMKIIalpha and beta-actin RNAs in low copy numbers. EMBO reports 12, 1077–1084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubing F. et al.. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci 30, 4160–4170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitelhofer M. et al.. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci 28, 7555–7562 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson J. H., Cui H. & Barbarese E. The balance of power in RNA trafficking. Curr Opin Neurobiol 11, 558–563 (2001). [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N. & Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004). [DOI] [PubMed] [Google Scholar]
- Brendza R. P., Serbus L. R., Duffy J. B. & Saxton W. M. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289, 2120–2122 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas J. N. et al.. Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A 105, 10820–10825 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas J. N. et al.. A role for huntington disease protein in dendritic RNA granules. J Biol Chem 285, 13142–13153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis E. H., Li Z., Singer R. H. & Taneja K. L. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol 123, 165–172 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook M. S., Lu M. & Kosik K. S. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci 20, 6385–6393 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J. & Li S. H. HAP1 and intracellular trafficking. Trends Pharmacol Sci 26, 1–3 (2005). [DOI] [PubMed] [Google Scholar]
- Twelvetrees A. E. et al.. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron 65, 53–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M., Zearfoss N. R. & Etkin L. D. Mechanisms of subcellular mRNA localization. Cell 108, 533–544 (2002). [DOI] [PubMed] [Google Scholar]
- Daigle N. & Ellenberg J. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat Methods 4, 633–636 (2007). [DOI] [PubMed] [Google Scholar]
- Stokin G. B. & Goldstein L. S. Axonal transport and Alzheimer's disease. Annu Rev Biochem 75, 607–627 (2006). [DOI] [PubMed] [Google Scholar]
- Gunawardena S. & Goldstein L. S. Polyglutamine diseases and transport problems: deadly traffic jams on neuronal highways. Arch Neurol 62, 46–51 (2005). [DOI] [PubMed] [Google Scholar]
- Harjes P. & Wanker E. E. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci 28, 425–433 (2003). [DOI] [PubMed] [Google Scholar]
- Eom T., Antar L. N., Singer R. H. & Bassell G. J. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci 23, 10433–10444 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E. et al.. Dynein light chain binding to a 3′-untranslated sequence mediates parathyroid hormone mRNA association with microtubules. J Clin Invest 105, 505–512 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. D. The molecular motor toolbox for intracellular transport. Cell 112, 467–480 (2003). [DOI] [PubMed] [Google Scholar]
- Colin E. et al.. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. Embo J 27, 2124–2134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her L. S. & Goldstein L. S. Enhanced sensitivity of striatal neurons to axonal transport defects induced by mutant huntingtin. J Neurosci 28, 13662–13672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda J. R. et al.. Genetic and pharmacological inhibition of calcineurin corrects the BDNF transport defect in Huntington's disease. Mol Brain 2, 33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoer S. R. et al.. Conventional kinesin holoenzymes are composed of heavy and light chain homodimers. Biochemistry 47, 4535–4543 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y. et al.. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. J Neurosci 30, 9349–9358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M. et al.. Phosphorylation of huntingtin at Ser421 in YAC128 neurons is associated with protection of YAC128 neurons from NMDA-mediated excitotoxicity and is modulated by PP1 and PP2A. J Neurosci 30, 14318–14329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. et al.. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell 137, 60–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge J. R. & Schmidt E. E. HAP1 can sequester a subset of TBP in cytoplasmic inclusions via specific interaction with the conserved TBP(CORE). BMC Mol Biol 8, 76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. J., Brown C. L., Maier K. C., Quintyne N. J. & Schroer T. A. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell 14, 5089–5097 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H. et al.. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J Biol Chem 279, 53427–53434 (2004). [DOI] [PubMed] [Google Scholar]
- Darzacq X. et al.. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173, 207–218 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. C., Freiman R. N., Goto H., Nishimoto T. & Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol 17, 6139–6146 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B. et al.. Localization of BDNF mRNA with the Huntington's disease protein in rat brain. Mol Neurodegener 5, 22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osten P. et al.. The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and alpha- and beta-SNAPs. Neuron 21, 99–110 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY INFORMATION
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Video 5
Supplementary Video 6


