Figure 2. Computational modeling framework for TRAIL signaling.
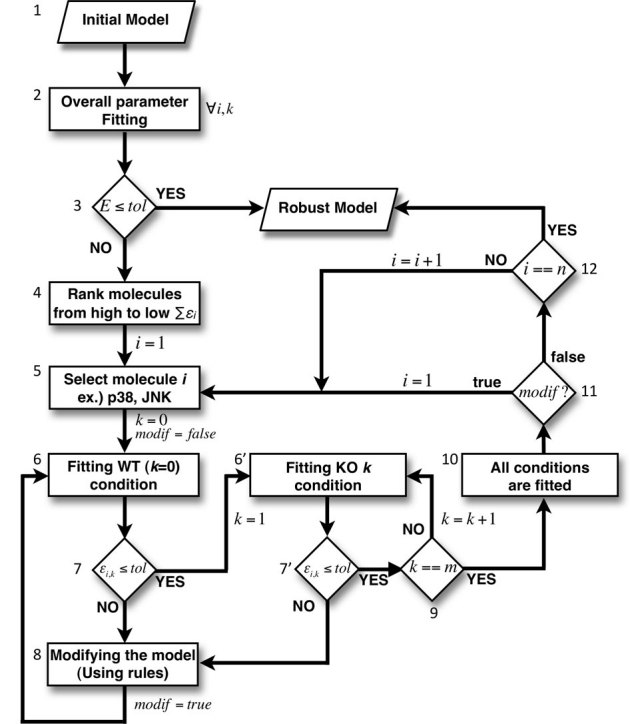
Parameters of the initial model based on the original topology (1) are determined by overall fitting of experimental data using a genetic algorithm (GA, see Methods) for all molecules (i = 1,2,..n) and at all conditions (k = 0,1,…m), here n = 5 and m = 4. (2). If the overall error E = max(ει,κ) between experimental and simulation profiles is higher than the set tolerance (3) (see Eq. 5) in Methods), the model is not acceptable. As the next step, the n molecules' activation profiles are ranked from the one showing highest (i = 1) to the lowest (i = n) error (4) for individual molecule's (5) best fitting in wildtype (6) and m other experimental conditions (6′). If the simulation of the ith molecule fit reasonably in the kth condition (individual error εi,k ≤ 0.15) (7), we check the next condition (k+1), else we modify the current topology according to response rules (8) (see Figure 3) and restart the procedure from wildtype condition again (6). If all m conditions fit for the ith molecule (9–10) without any changes applied to the topology, we proceed to the next molecule (i+1) (11). If any change is necessary to the topology, the parameters have to be refitted for all molecules from the first molecule (i = 1). The whole procedure is repeated until the resultant model fit all experimental profiles of the n molecules within the error tolerance (12).
