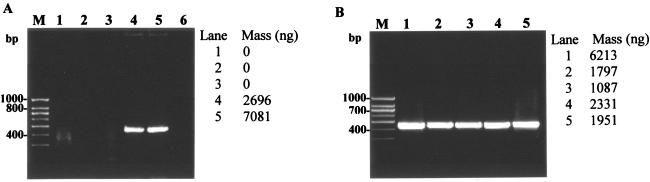
FIG. 1.
Optimization of cell lysis protocol for direct PCR-based detection of mycobacteria. (A) Optimization of chemical reagent for cell lysis as a prestep in hsp-based PCR amplification of M. smegmatis. Lanes 1 to 5, comparison of five different lysis reagents (reagents 1 through 5) containing various concentrations of SDS and Triton X-100 in Tris-EDTA buffer used in combination with thermal regime I (Table 1); lane 6, negative control (no template DNA). (B) Optimization of heating regimes for cell lysis of M. smegmatis using lysis reagent 5 (Table 1) in the hsp-based PCR. Lanes 1 to 5, comparison of thermal regimes 1 through 5 (Table 2); PCR amplification was based on mycobacterium-specific hsp gene with an expected 439-bp amplicon as described in the text. Lane M, 100-bp DNA size marker (PGC Scientifics).