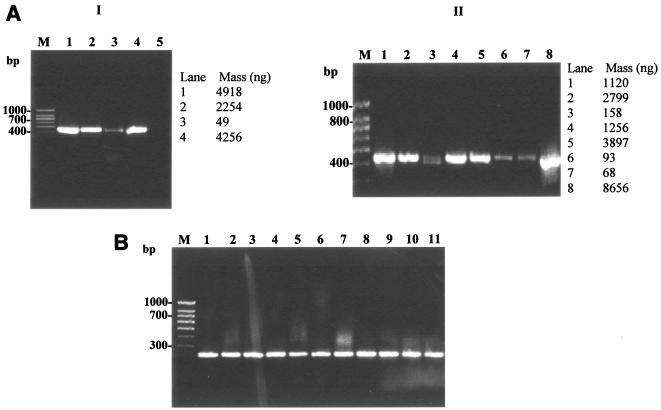
FIG. 2.
Evaluation of the optimized protocols for cell lysis and hsp-based PCR for mycobacteria. (A) Evaluation of the optimized cell lysis protocol (lysis reagent 5, thermal regime V) for hsp-based PCR amplification (439 bp) of mycobacterial reference strains and MWF isolates. Panel I: lanes 1 to 4, M. smegmatis, M. chelonae, M. immunogenum, and M. bovis; lane 5, no-template control. Panel II: lanes 1 to 8, M. smegmatis, M. chelonae, M. immunogenum, and Mycobacterium isolates M-JY1, M-JY2, M-JY3, M-JY4 and M-JY5; lane M, 100-bp DNA size marker (PGC Scientifics). PCR primers and conditions were the same as described in the legend of Fig. 1. (B) Evaluation of new hsp-based PCR primers and modified PCR conditions for genus-specific PCR amplification (228 bp) of mycobacteria. Lanes 1 to 3, M. smegmatis, M. chelonae, and M. immunogenum; lanes 4 to 11, Mycobacterium isolates M-JY1, M-JY2, M-JY3, M-JY4, M-JY5, M-JY6, M-JY7 and M-JY8; lane M, 100-bp DNA size marker (PGC Scientifics). Cells were lysed using the optimized direct cell lysis method as described for panel A, and the lysates were amplified using new PCR primers and conditions described in the text for amplification of a 228-bp PCR product of the hsp gene.