Abstract
One hundred eighty-four clinical isolates of Klebsiella pneumoniae were recovered from August 1996 to October 1997 at the Pediatric Hospital of the Instituto Mexicano del Seguro Social in Mexico City, Mexico. Most of the isolates were collected from the neonatal intensive care unit and infant wards, which are located on the same floor of the hospital. Isolates were genotypically compared by pulsed-field gel electrophoresis with XbaI restriction of chromosomal DNA. Of 184 clinical isolates, 91 belonged to cluster A and comprised three subtypes (A1, A2, and A3), while 93 isolates, comprising two minor clones, B (10 isolates) and C (7 isolates), and 76 unique patterns, were considered unrelated isolates (URI). Susceptibility patterns were indistinguishable in both groups. Fifty extended-spectrum β-lactamase-producing isolates, including 34 from clone A and 16 from URI, were examined for further studies. Molecular and genetic analysis showed that 47 of 50 clinical isolates expressed the SHV-5 β-lactamase. This enzyme, in combination with TEM-1, was encoded in a ≥170-kb conjugative plasmid. Results indicate that dissemination of this resistance was due to clonal and horizontal spread.
Klebsiella spp. have been prominent among gram-negative bacilli causing nosocomial infections, as well as being an important source of transferable antibiotic resistance. During the 1970s there were frequent epidemics of gentamicin-resistant Klebsiella pneumoniae infections in hospitals (5). In recent years, following the overuse of expanded-spectrum cephalospirins, several outbreaks caused by extended-spectrum β-lactamases (ESBLs) have been reported (5, 18, 20, 23). ESBLs are enzymes with considerable hydrolyzing activity on a wide variety of β-lactam antibiotics, including oxyaminocephalosporins and aztreonam (12). Such enzymes have been shown to be derived from SHV- or TEM-type β-lactamases by one or more amino acid substitutions (11, 21). Since the first ESBL-expressing isolate was discovered, many types of ESBLs, exhibiting high degrees of diversity in their structures and activities, have been characterized and described. Several families reflecting evolutionary and/or functional similarities can be distinguished (19). Among the most prevalent types of ESBLs are members of the TEM and SHV families (3). To date about 17 amino acid positions in ESBL protein sequences have been reported to be heterogeneous (7). Isolation and sequencing of natural genes encoding ESBLs provide important data from an epidemiological point of view and contribute to our understanding of the structure and function of β-lactamases. The genes coding for ESBLs are usually carried by plasmids, which strongly facilitate their spread among strains of many species of gram-negative bacteria. Nevertheless, K. pneumoniae and Escherichia coli remain the most frequently reported ESBL producers (18, 20, 22, 23).
There is limited information in Mexico concerning molecular studies on the type of ESBL selected in vivo in multidrug-resistant enterobacteria (13, 24, 25, 26). In this work we used a molecular approach to determine the epidemiology of an outbreak produced by an endemic multidrug-resistant K. pneumoniae strain.
MATERIALS AND METHODS
Hospital setting.
The study was carried out in a tertiary-care pediatric hospital in Mexico City with 194 beds and five areas of hospitalization: (i) a pediatric intensive-care unit (ICU) with 18 beds; (ii) an area for preschool children, with 40 beds; (iii) a neonatal ICU (NICU) with 24 beds; (iv) an area for infants, with 52 beds; and (v) an area for school-age children and adolescents, with 60 beds. All patients are referred from seven hospitals in Mexico City and from four Mexican states (Morelos, Guerrero, Querétaro, and Chiapas).
Bacterial strains.
One hundred eighty-four clinical isolates of K. pneumoniae were isolated from August 1996 to November 1997. During this period an increase in the incidence of isolation was detected. The species of the organisms were verified by tests with the API 20E system (bioMerieux, Marcy L′Etoile, France). Genetic and molecular characterization studies for ESBL production were performed on 50 clinical isolates selected according to their pulsed-field gel electrophoresis (PFGE) patterns: 21 corresponding to clone A; 11 corresponding to subtype A1; 1 isolate each for subtype A2, subtype A3, clone B, and clone C; and 14 with unique patterns (UNP). All strains were isolated from blood or cerebrospinal fluid (CSF) cultures (see Table 2).
TABLE 2.
Epidemiological and molecular characteristics of K. pneumoniae ESBL-producing clinical isolates
| Strain no. | Date of isolation (day/mo/yr) | Ward | Sample | PFGE pattern | β-Lactamase pattern (pl)a | Conjugation | Resistance pattern of transconjugantb |
|---|---|---|---|---|---|---|---|
| 1533A | 16/8/1996 | School-age | Blood | A | 5.4, 7.3, (8.2) | + | a |
| 1545A | 12/9/1996 | School-age | Blood | A | 5.4, 7.3, (8.2) | + | b |
| 1607A | 12/11/1996 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | d |
| 1517A | 18/2/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | b |
| 1589A | 10/4/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | b |
| 1591A | 8/5/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | b |
| 1594A | 26/5/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | c |
| 1596A | 8/7/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | − | |
| 1599A | 15/7/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | a |
| 1598A | 15/7/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | c |
| 1610A | 21/7/1997 | Infant | Blood | A | 5.4, 7.3, (8.2) | + | c |
| 1602A | 8/8/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | − | |
| 1604A | 20/8/1997 | Infant | Blood | A | 5.4, 7.3, (8.2) | + | d |
| 1606A | 25/8/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | c |
| 1611A | 8/9/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | − | |
| 1620A | 29/9/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | a |
| 1625A | 30/9/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | d |
| 1516A | 31/1/1997 | NICU | Blood | A | 5.4, 7.3, (8.2) | + | d |
| 1525A1 | 15/8/1996 | NICU | Blood | A1 | 5.4, 7.3, (8.2) | − | |
| 1547A1 | 12/9/1996 | NICU | Blood | A1 | 5.4, 7.3, (8.2) | + | b |
| 1574A1 | 11/11/1996 | NICU | Blood | A1 | 5.4, 7.3, (8.2) | − | |
| 1510A1 | 27/12/1996 | School-age | Blood | A1 | 5.4, 7.3, (8.2) | + | d |
| 1513A1 | 3/1/1997 | NICU | Blood | A1 | 5.4, 7.3, (8.2) | + | b |
| 1583A1 | 21/1/1997 | NICU | Blood | A1 | 5.4, 7.3, (8.2) | + | b |
| 1582A1 | 27/1/1997 | NICU | Blood | A1 | 5.4, 7.3, (8.2) | + | b |
| 1584A1 | 30/1/1997 | Infant | Blood | A1 | 5.4, 7.3, (8.2) | + | b |
| 1515A1 | 31/1/1997 | NICU | CSF | A1 | 5.4, 7.3, (8.2) | + | b |
| 1585A1 | 19/3/1997 | NICU | CSF | A1 | 5.4, 7.3, (8.2) | + | b |
| 1523A1 | 2/4/1997 | NICU | CSF | A1 | 5.4, 7.3, (8.2) | + | c |
| 1511A3 | 2/1/1997 | NICU | Blood | A3 | 5.4, 7.3, (8.2) | + | b |
| 1528UNP | 23/8/1996 | Infant | Blood | UNP | 5.4, 7.3, (8.2) | + | a |
| 1549UNP | 17/9/1996 | Infant | Blood | UNP | 5.4, 7.3, (8.2) | + | b |
| 1608UNP | 19/11/1996 | NICU | Blood | UNP | 5.4, 7.3, (8.2) | + | d |
| 1578B | 25/11/1996 | Infant | Blood | B | 5.4, 7.3, (8.2) | + | b |
| 1603UNP | 11/6/1997 | NICU | Blood | UNP | 5.4, 7.3, (8.2) | − | |
| 1621UNP | 26/9/1997 | NICU | Blood | UNP | 5.4, 7.3, (8.2) | + | c |
| 1628UNP | 3/10/1997 | NICU | Blood | UNP | 5.4, 7.3, (8.2) | + | c |
| 1627UNP | 6/10/1997 | School-age | Blood | UNP | 5.4, 7.3, (8.2) | + | b |
| 1521A | 17/3/1997 | NICU | Blood | A | 5.4, 7.0, (8.2) | + | b |
| 1560UNP | 25/9/1996 | Infant | Blood | UNP | 5.4, 7.0, (8.2) | + | a |
| 1563UNP | 25/9/1996 | Infant | Blood | UNP | 5.4, 7.0, (8.2) | + | a |
| 1600UNP | 4/8/1997 | School-age | Blood | UNP | 5.4, 7.0, (8.2) | + | b |
| 1509A | 20/12/1996 | NICU | Blood | A | 5.4, (8.2) | + | a |
| 1542UNP | 8/8/1996 | NICU | Blood | UNP | 5.4, (8.2) | + | a |
| 1568C | 2/10/1996 | Infant | CSF | C | 5.4, (8.2) | − | |
| 1601UNP | 4/8/1997 | Outpatient | CSF | UNP | 5.4, (8.2) | + | c |
| 1623UNP | 1/10/1997 | Infant | Blood | UNP | 5.4, (8.2) | + | b |
| 1529A | 26/8/1996 | NICU | Blood | A | 5.4, (7.6) | + | a |
| 1520UNP | 4/3/1997 | Emergency | Blood | UNP | 5.4, (7.6) | + | a |
| 1562A2 | 30/9/1996 | Infant | CSF | A2 | 5.4, (7.6) | − |
A pI in parentheses indicates that the enzyme is an (ESBL), as detected by bioassay. Boldfaced data are values observed for transconjugants.
a, resistant to ampicillin, cefotaxime, kanamycin, tetracycline, chloramphenicol, and gentamicin; b, resistant to all antibiotics except tetracycline; c, resistant to all antibiotics except chloramphenicol; d, resistant to all antibiotics except tetracycline and chloramphenicol.
Antimicrobial agents.
Pure salts of the following drugs were provided by the companies indicated: cefotaxime and cefpirome (Hoechst-Marion-Roussel, Romainville, France), ceftazidime (GlaxoWellcome, Mexico City, Mexico), aztreonam and cefepime (Bristol-Myers Squibb, Mexico City, Mexico), and clavulanic acid (SmithKline Beecham Pharmaceuticals, Mexico City, Mexico). Rifampin, tetracycline. and gentamicin were obtained from Sigma (St. Louis, Mo.).
Susceptibility testing and ESBL test.
MICs were determined by agar dilution on Mueller-Hinton agar according to the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS). Breakpoints for resistance (in micrograms per milliliter) were as follows: ampicillin, ≥32; carbenicillin, ≥64; chloramphenicol, ≥32; amikacin, ≥64; gentamicin, ≥16; ceftazidime, ≥32; cefotaxime, ≥64; ceftizoxime, ≥64; cefepime, ≥32; imipenem, ≥16; meropenem, ≥16; norfloxacin, ≥16. ESBL production was performed by the disk diffusion method with cefotaxime and ceftazidime alone and in combination with clavulanic acid, as recommended by the NCCLS (17).
Genome fingerprinting by PFGE.
For PFGE typing, whole-cell DNA was obtained according to the method of Miranda et al. (16). DNA was digested with the XbaI restriction enzyme (Gibco BRL, Gaithersburg, Md.) and separated in a 1% agarose gel (Pulsed Field-Certified; Pronadisa, Madrid, Spain) with a Gene-Path System (Bio-Rad, Hercules, Calif.). The gel was stained with ethidium bromide and visualized with the Gel-Doc system (Bio-Rad). The images were interpreted with Multi-analyst software (Bio-Rad) according to the criteria of Tenover et al. (28).
IEF and bioassay.
Isoelectric focusing (IEF) was conducted according to the method described by Matthew et al. (14) by using a Phast system minigel with a pH range of 3 to10 (Pharmacia, LKB). Extracts from TEM-1-, SHV-2-, and SHV-5-producing strains were used as standards for pIs of 5.4, 7.6, and 8.2, respectively. To determine the ESBLs encoded by the strains, a bioassay was performed as described by Silva-Sanchez and Aguilar-Zacarias (27).
Plasmid isolation and conjugation experiments.
DNA was extracted from clinical isolates and transconjugants according to the method described by Kieser (10). DNA was visualized after vertical electrophoresis in 0.7% agarose gels stained with ethidium bromide. Plasmids R6K (40 kb), RP4 (5 kb), IR (93 kb), and pUD21 (170 kb) were used as molecular weight markers. Matings were performed as described by Miller (15), by using E. coli strain J53-2 (F− pro met Rifr). In all cases, transconjugants were selected on Luria agar supplemented with rifampin (200 μg/ml) in combination with cefotaxime (1 μg/ml) or ampicillin (100 μg/ml). For each successful mating experiment, 25 independent transconjugants were obtained from each selection medium and were tested on Luria plates supplemented with ampicillin (100 μg/ml), cefotaxime (1 μg/ml), kanamycin (25 μg/ml), tetracycline (25 μg/ml), chloramphenicol (10 μg/ml), and gentamicin (1 μg/ml).
TEM- or SHV-specific PCR and DNA sequencing.
To amplify TEM-related genes from clinical isolates, oligonucleotide primers OT1 and OT2, described by Arlet and Philippon (1), were used for PCR. For SHV-specific PCR, primers SE5 and SB3 were used as described by Silva et al. (26). The PCR mixture for both amplifications (50 μl) contained 30 pmol of each primer, 300 ng of total DNA, 1× reaction buffer, 200 μM MgSO2, 200 μM each deoxynucleoside triphosphate, and 2.5 U of Taq DNA polymerase. Enhancer buffer (Gibco BRL) at a final concentration of 3× was added to the reaction mixture to enhance the specificity of hybridization. The PCR amplification conditions for both reactions were as follows: initial denaturation (95°C for 5 min); 30 cycles of denaturation (94°C for 30s), annealing (58°C for 30s), and polymerization (72°C for 120s); and an additional polymerization step (72°C for 15 min) at the end of the program. The product of blaSHV amplification was used to determine the nucleotide sequence with the fluorescence-based Taq FS Dye terminator cycle sequencing kit and the same primers. Sequence analysis was performed with Genetics Computer Group software and by BLASTx searching (of the EMBL, SwissProt, and PIR databases). Multiple alignment was performed with the Clustal W program (29).
Nucleotide sequence accession numbers.
The nucleotide sequences of shv genes from strains X1529A, X1620A, X1623UNP, R1549UNP, and X1600UNP have been deposited in GenBank under accession no. AY386365 to AY386369, respectively.
RESULTS
Genomic typing of K. pneumoniae isolates.
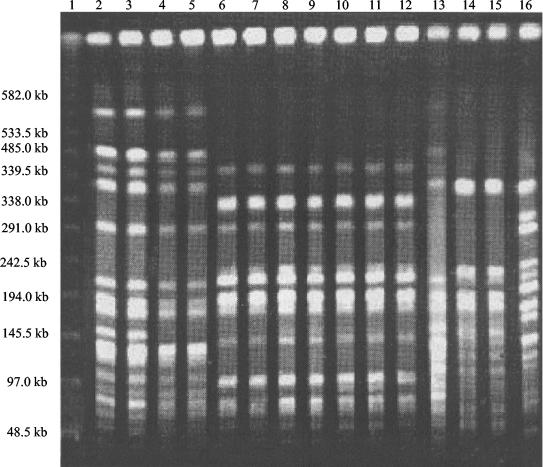
Between August 1996 and October 1997, 184 clinical isolates of K. pneumoniae were collected and typed by PFGE. Six different major PFGE types were identified. Most of the isolates corresponded to the major type (clone A), which included 91 clinical isolates (49.5%), suggesting the presence of one predominant clone of K. pneumoniae. The other 93 isolates (50.5%) comprised two minor groups, clone B (10 isolates) and clone C (7 isolates), and 76 UNP; all of these were referred as unrelated isolates (URI). Isolates from clone A could be further classified into subtypes A1, A2, and A3. A representative sample of the patterns is shown in Fig. 1. Most clinical isolates were obtained from patients in the NICU and the infant ward, both located on the same floor of the hospital. In order to perform further genetic and molecular characterizations, 50 strains of the 184 clinical isolates were selected, including 21 from clone A, 11 from subtype A1, 14 with UNP, and 4 isolates corresponding, respectively, to pattern A2, A3, B, or C. All 50 clinical isolates were identified as ESBL producers.
FIG. 1.
Representative agarose gel PFGE of XbaI-digested genomic DNA of ESBL-producing K. pneumoniae isolates. Lane 1, molecular size marker of lambda ladder; lanes 2 to 5 and 13, clone B; lanes 6 to 12, clone A; lanes 14 and 15, clone C; lane 16, UNP.
Antibiotic susceptibilities.
MICs of various antibiotics were determined by agar dilution against the 184 K. pneumoniae clinical isolates. MICs at which 50 and 90% of the isolates were inhibited (MIC50 and MIC90, respectively) are given in Table 1. All strains were resistant to β-lactam antibiotics and aminoglycosides and were susceptible to imipenem, meropenem, and norfloxacin. There was no difference in MICs between the clone A isolates and URI isolates.
TABLE 1.
Comparison of antimicrobial susceptibilities between K. pneumoniae clone A isolates and URI
| Antibiotic | MIC (μg/ml)a for:
|
|||
|---|---|---|---|---|
| Clone A
|
URIb
|
|||
| 50% | 90% | 50% | 90% | |
| Ampicillin | >64 | >64 | >64 | >64 |
| Carbenicillin | >256 | >256 | >256 | >256 |
| Chloramphenicol | >16 | >16 | >16 | >16 |
| Amikacin | >64 | >64 | 16 | >64 |
| Gentamicin | >64 | >64 | 64 | >64 |
| Ceftazidime | >64 | >64 | 64 | >64 |
| Cefotaxime | <16 | >64 | <16 | >64 |
| Ceftizoxime | <8 | 64 | >8 | >64 |
| Cefepime | <4 | 16 | <4 | 32 |
| Imipenem | <4 | <4 | <4 | <4 |
| Meropenem | <4 | <4 | <4 | <4 |
| Norfloxacin | <2 | 2 | <2 | 2 |
50% and 90%, MIC50 and MIC90.
Include clinical isolates of clone B, clone C, and UNP.
Plasmid profiles and resistance transfer.
All 50 clinical isolates harbored at least two plasmids, one of ≥170 kb and one of 40 to 90 kb. To identify the plasmid responsible for conferring cefotaxime resistance, all clinical isolates of K. pneumoniae were mated to a susceptible E. coli host. Transconjugants were selected on ampicillin or cefotaxime in independent selections and were designated with the letter X and the number of the parental strain. Successful matings were detected for 42 of the 50 clinical isolates; of these, 18 corresponded to clone A, 9 to subtype A1, 1 to subtype A3, 1 to clone B, and 13 to UNP. Ampicillin resistance transfer was successful in all 42 matings; however, when selection was done on cefotaxime, only 27 of 42 transconjugants (64.3%) were obtained. However, the transconjugants that grew only on ampicillin showed cefotaxime resistance in a later test. Frequencies of resistance transfer were 10−2 to 10−5 transconjugants per donor cell for ampicillin and 10−4 to 10−6 for cefotaxime. In general, resistance transfer frequencies were one- to twofold higher for ampicillin than for cefotaxime. All 42 transconjugants harbored only the largest plasmid (≥170 kb) and expressed resistance markers for other antibiotics such as tetracycline, kanamycin, gentamicin, and chloramphenicol. Eighteen transconjugantswere susceptible only to tetracycline, 8 were susceptible only to chloramphenicol, 6 were susceptible only to tetracycline and chloramphenicol, and 10 were resistant to the six antibiotics tested.
IEF analysis and enzyme inhibition test.
Crude sonicates of the 50 clinical isolates and 42 transconjugants were subjected to IEF in order to identify the β-lactamase profiles. All clinical isolates and transconjugants expressed a β-lactamase with a pI of 5.4 and two or three additional bands with pIs of 7.0, 7.3, 7.6, or 8.2 (Table 2). One predominant β-lactamase profile with pIs of 5.4, 7.3, and 8.2 was identified for 38 clinical isolates, of which 18 corresponded to clone A, 11 to subtype A1, 1 to subtype A3, 1 to clone B, and 7 to UNP. Three other, minor β-lactamase profiles were identified for clinical isolates: the first with pIs of 5.4, 7.0, and 8.2, the second with pIs of 5.4 and 8.2, and the last with pIs of 5.4 and 7.6. In all cases the last enzyme, with a pI of 8.2 or 7.6, was determined to have the capacity to hydrolyze cefotaxime in the bioassay, indicating correspondence to an ESBL. When β-lactamase profiles were determined for transconjugants, a major pattern of enzymes with pIs of 5.4 and 8.2 was identified. This pattern was expressed in 40 of 42 transconjugants, of which 17 corresponded to clone A, 9 to subtype A1, 1 to subtype A3, 1 to clone B, and 12 to UNP. Two transconjugants, 1 from clone A and 1 with a UNP, showed a β-lactamase pattern with pIs of 5.4 and 7.6.
PCR amplification of blaTEM and blaSHV genes.
According to the pIs of the β-lactamases obtained, the enzyme with a pI of 5.4 should correspond to the TEM-1 β-lactamase, and enzymes with a pI of 7.6 or 8.2 should correspond to SHV-derived β-lactamases. In order to corroborate this hypothesis, PCR assays were performed with specific oligonucleotides. Total-DNA preparations of five different clinical isolates and the respective transconjugants, representing the four β-lactamase profiles, were tested for the presence of blaTEM and blaSHV genes. In the first case, an amplification product of the expected size (503 bp) was obtained for all clinical isolates and transconjugants tested, indicating that the β-lactamase with a pI of 5.4 corresponded to TEM-1 (Table 3). When PCR amplification was performed for the detection of blaSHV genes, a product of approximately 900 bp was observed in all cases.
TABLE 3.
Comparison of amino acid sequences of SHV derivatives and detection of TEM β-lactamase in K. pneumoniae clinical isolates and transconjugants
| Strain no.a | β-Lactamase pattern (pl)b | TEMc | ESBL | Amino acid at position:
|
Source or reference | GenBank accession no. | |
|---|---|---|---|---|---|---|---|
| 238 | 240 | ||||||
| (7.6) | − | SHV-1 | G | E | 7 | AF148850 | |
| (7.6) | − | SHV-2 | S | E | 7 | AF148851 | |
| (8.2) | − | SHV-5 | S | K | 7 | X55640 | |
| X1529A | 5.4, (7.6) | + | SHV-2 | S | E | This work | AY386365 |
| X1620A | 5.4, (8.2) | + | SHV-5 | S | K | This work | AY386366 |
| X1623UNP | 5.4, (8.2) | + | SHV-5 | S | K | This work | AY386367 |
| R1549UNP | 5.4, 7.3, (8.2) | + | SHV-5 | S | K | This work | AY386368 |
| X1600UNP | 5.4, 7.0, (8.2) | + | SHV-5 | S | K | This work | AY386369 |
R, clinical isolate; X, transconjugant.
A pI in parentheses indicates an ESBL, as detected by a bioassay.
Presence (+) or absence (−) as determined by PCR.
Sequencing of ESBL-encoding genes.
The products of PCR amplification of the blaSHV genes from E. coli transconjugants X1600UNP, X1620A, and X1623UNP and from clinical isolate R1549A, all of which expressed an enzyme with a pI of 8.2, were used for sequencing. The results were compared with the sequence of the blaSHV-1 gene and indicated that all contained the Gly238Ser and Glu240Lys substitutions, which correspond to the SHV-5 β-lactamase. Meanwhile, the DNA sequence of the PCR product from E. coli transconjugant X1529A, producing the enzyme with a pI of 7.6, encoded only the Gly238Ser amino acid substitution, corresponding to the SHV-2 gene (Table 3).
DISCUSSION
A total of 184 clinical isolates of K. pneumoniae were recovered between August 1996 and October 1997 at the Pediatric Hospital of the Instituto Mexicano del Seguro Social in Mexico City. Most of the isolates were recovered from the NICU and the infant ward, which are located on the same floor. According to the genotyping, a single clonal spread was detected by PFGE in 50% of the isolates while the other 50% comprised URI, which included two minor clones (B and C) and UNP. Both groups harbor a multidrug resistance plasmid. Therefore, this outbreak was due to the spread of an epidemic strain of multidrug-resistant K. pneumoniae (2, 5, 8, 26). At the same time, it was due to the dissemination of a multidrug resistance plasmid to other clinical isolates, as has occurred in other hospitals (13, 23).
Recent studies of hospital-associated infections in the United States have reported that SHV-4 and SHV-5 are becoming the predominant types of ESBLs found in nosocomial isolates of K. pneumoniae. In Germany, SHV-2 and SHV-5 seem to be predominant; and in France, SHV-3, SHV-4, and TEM-3 are more common. SHV-2 is widespread internationally (9).
In order to characterize cefotaxime resistance at the molecular level, 50 clinical isolates were randomly selected; they included type A, with three subtypes, and URI, which included two minor clones (B and C) and UNP. In all clinical isolates, TEM-1 and SHV-5 (and in a minor proportion, SHV-2) enzymes were encoded in a self-transferable ≥170-kb plasmid. In addition to these two enzymes, a third, non-ESBL β-lactamase with a pI of 7.0 or 7.3 was expressed. This β-lactamase was not transferred by conjugation to the E. coli recipient strain, suggesting that this enzyme is not carried on the same plasmid. The prevalence of ESBL producers was almost the same throughout the study period, suggesting that SHV-type β-lactamase-producing strains were already endemic in the hospital.
The identification of SHV-2 and SHV-5 makes it possible to speculate on the evolutionary sequence of SHV-type ESBLs, i.e., from SHV-2 to SHV-5, according to the mutation process. Because only one amino acid substitution (Gly240Lys) is required, it may be assumed that in this hospital the mutation process in K. pneumoniae clinical isolates was sequential from SHV-2 to SHV-5, thereby disseminating to other strains and/or patients.
When the mating experiments were developed in both genotyping groups (A and URI), the results were very similar, 82 and 88%, respectively. Two antibiotics were used independently for selection, ampicillin and cefotaxime, and frequencies of transfer of resistance to ampicillin were always one- to twofold higher than those for cefotaxime. Considering that the ≥170-kb plasmid encodes two β-lactamases, TEM-1 and SHV-2 or SHV-5, these results suggest that the higher frequency for ampicillin resistance transfer may have been due to the fact that TEM-1 expresses preferentially to the ESBL enzyme, conferring resistance to ampicillin in E. coli transconjugants. The cefotaxime phenotype depends on the expression of the SHV-derived enzyme. Also, variations in non-β-lactam resistance markers associated with the ≥170-kb plasmid suggest the ability of the plasmid to accept or lose these genes. These could be included in other mobile genetic elements such as transposons or integrons (4). Also, the fact that a minor number of strains were unable to transfer resistance could be due to loss of conjugation by means of a possible mutation along the tra operon, which showed diversity in the clinical isolates. In these isolates, the high frequency of conjugation increases the possibility of dissemination of multidrug resistance among other genera in vivo.
Other studies investigating β-lactamases and ESBL types in different clinical isolates from Mexico, Poland, and Taiwan (6, 13, 26) have documented the SHV-5 enzyme in association with the TEM-1 β-lactamase. In the future, our laboratory will undertake investigations using molecular biology techniques to determine whether the plasmids expressing these enzymes are related at the molecular level.
On the basis of susceptibilities to antibiotics, genotyping, β-lactamase production, and conjugation experiments, it is possible that spread of an endemic strain and horizontal gene transfer were responsible for the high frequency of detection of K. pneumoniae ESBL producers in this setting. Most of these strains were obtained from the NICU and the infant ward, indicating a localized dissemination within the hospital and pointing to a potential source of spread of an SHV-5 ESBL-encoding plasmid in the hospital.
In conclusion, this study highlights the need to establish an antimicrobial resistance surveillance network for K. pneumoniae to monitor the trends and new types of resistance mechanisms in this hospital. Also, the factors responsible for the selection and dissemination of this plasmid encoding the SHV-derived enzyme and clone A need to be identified, controlled, and, where possible, prevented so as to avoid major outbreaks.
Acknowledgments
This work was supported by grants 28726-M and 30938 from CONACYT.
We thank Zita Becerra for excellent laboratory assistance.
REFERENCES
- 1.Arlet, G., and A. Philippon. 1991. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett. 66:19-25. [DOI] [PubMed] [Google Scholar]
- 2.Arlet, G., M. Rouveau, G. Fournier, P. H. Lagrange, and A. Philippon. 1993. Novel, plasmid-encoded, TEM-derived extended-spectrum beta-lactamase in Klebsiella pneumoniae conferring higher resistance to aztreonam than to extended-spectrum cephalosporins. Antimicrob. Agents Chemother. 37:2020-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 5.French, G. L., K. P. Shannon, and N. Simmons. 1996. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and β-lactam-β-lactamase-inhibitor combinations by hyperproduction of SHV-5 β-lactamase. J. Clin. Microbiol. 34:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gniadkowski, M., I. Schneider, R. Jungwirth, W. Hryniewicz, and A. Bauernfeind. 1998. Ceftazidime-resistant Enterobacteriaceae isolates from three Polish hospitals: identification of three novel TEM- and SHV-5-type extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 42:514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G. A., and K. Bush. 5June2003, posting date. Amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant β-lactamases.[Online.] Lahey Clinic, Burlington, Mass. http://www.lahey.org/Studies/?D=http://www.lahey.org/studies/webt.stm&C=404.
- 8.Jacoby, G. A., and P. Han. 1996. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J. Clin. Microbiol. 34:908-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby, G. A., and A. A. Medeiros. 1991. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:1697-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 11.Knox, J. R., P. C. Moews, and J. M. Frere. 1996. Molecular evolution of bacterial beta-lactam resistance. Chem. Biol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 12.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Aguilar, G., C. M. Alpuche-Aranda, C. Anaya, D. Alcantar-Curiel, C. Gayosso, C. Daza, C. Mijares, J. C. Tinoco, and J. I. Santos. 2001. Outbreak of nosocomial sepsis and pneumonia in a newborn intensive care unit by multiresistant extended-spectrum beta-lactamase-producing Klebsiella pneumoniae: high impact on mortality. Infect. Control Hosp. Epidemiol. 22:725-728. [DOI] [PubMed] [Google Scholar]
- 14.Matthew, M., R. W. Hedges, and J. T. Smith. 1979. Types of β-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 138:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1992. Experiments in molecular genetics, p. 82-85. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Miranda, G., C. Kelly, F. Solorzano, B. Leanos, R. Coria, and J. E. Patterson. 1996. Use of pulsed-field gel electrophoresis typing to study an outbreak of infection due to Serratia marcescens in a neonatal intensive care unit. J. Clin. Microbiol. 34:3138-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2002. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Nouvellon, M., J. L. Pons, D. Sirot, M. L. Combe, and J. F. Lemeland. 1994. Clonal outbreaks of extended-spectrum β-lactamase-producing strains of Klebsiella pneumoniae demonstrated by antibiotic susceptibility testing, β-lactamase typing, and multilocus enzyme electrophoresis. J. Clin. Microbiol. 32:2625-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palzkill, T. 1998. β-Lactamases are changing their activity spectrums. ASM News 64:90-95. [Google Scholar]
- 20.Peña, C., M. Pujol, C. Ardanuy, A. Ricart, R. Pallares, J. Linares, J. Ariza, and F. Gudiol. 1998. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 42:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitout, J. D., C. C. Sanders, and W. E. Sanders, Jr. 1997. Antimicrobial resistance with focus on beta-lactam resistance in gram-negative bacilli. Am. J. Med. 103:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prodinger, W. M., M. Fille, A. Bauernfeind, I. Stemplinger, S. Amann, B. Pfausler, C. Lass-Florl, and M. P. Dierich. 1996. Molecular epidemiology of Klebsiella pneumoniae producing SHV-5 β-lactamase: parallel outbreaks due to multiple plasmid transfer. J. Clin. Microbiol. 34:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva, J., C. Aguilar, G. Ayala, M. A. Estrada, U. Garza-Ramos, R. Lara-Lemus, and L. Ledezma. 2000. TLA-1: a new plasmid-mediated extended-spectrum β-lactamase from Escherichia coli. Antimicrob. Agents Chemother. 44:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva, J., C. Aguilar, Z. Becerra, F. Lopez-Antunano, and R. Garcia. 1999. Extended-spectrum β-lactamases in clinical isolates of enterobacteria in Mexico. Microb. Drug Resist. 5:189-193. [DOI] [PubMed] [Google Scholar]
- 26.Silva, J., R. Gatica, C. Aguilar, Z. Becerra, U. Garza-Ramos, M. Velazquez, G. Miranda, B. Leanos, F. Solorzano, and G. Echaniz. 2001. Outbreak of infection with extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a Mexican hospital. J. Clin. Microbiol. 39:3193-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva-Sanchez, J., and C. Aguilar-Zacarias. 1997. β-Lactamase bioassay: a simplified method to determine extended-spectrum β-lactamases (ESBL) in enterobacteria. Arch. Med. Res. 28:285-287. [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]