Abstract
Delta ligands are important for regulating Notch signaling through transcellular stimulation of Notch receptors. The cytoplasmic tails of Delta ligands have multiple potential regulatory sites including several lysine residues that are putative targets for ubiquitination by the E3 ubiquitin ligases, Mind Bomb and Neuralized. To identify possible roles for specific lysine residues in the cytoplasmic tail of the Notch ligand Dll1 a mutational and functional analysis was performed. Examination of a panel of individual or clustered lysine mutants demonstrated that lysine 613 (K613) in the cytoplasmic tail of Dll1 is a key residue necessary for transcellular activation of Notch signaling. Multi-ubiquitination of the Dll1 mutant Dll1-K613R was altered compared to wild type Dll1, and the K613R mutation blocked the ability of Dll1 to interact with Notch1. Finally, mutation of K613 did not affect the stability of Dll1 or its ability to traffic to recycle to the plasma membrane, but did enhance the fraction associated with lipid rafts. Collectively these results suggest that the transcellular defect in Notch signaling attributed to residue K613 in cytoplasmic tail of Dll1 may result from altering its multi-ubiquitination and increasing its retention in lipid rafts.
Keywords: Notch, Delta, ubiquitin, endocytosis, recycling, lipid raft microdomains
1.0 Introduction
The Notch signaling pathway plays important roles in specifying cell fate during embryonic development and contributes to adult homeostasis[1, 2]. Notch signaling is activated by transcellular interaction between ligands of the Delta/Serrate/Lag2 (DSL) family in signal-sending cells with Notch receptors in signal-receiving cells. Five DSL ligands have been identified in mammals including three Delta-like proteins (Dll1, 3, and 4) and two Jagged (Jagged1 and Jagged2) proteins[3, 4]. Notch receptors appear on the plasma membrane of cells as non-covalently bound heterodimers that are generated from a 300 kDa precursor form by proteolytic cleavage (S1) mediated by a furin-convertase in the trans-Golgi. The cleaved Notch1 heterodimer is composed of a 120 kDa Notch intracellular domain (Notch-ICD) subunit bound to its 180 kDa Notch extracellular domain subunit (Notch-ECD)[5–7]. In response to transcellular binding between Notch and a member of the DSL ligand family, a second protease cleavage (S2) site is exposed and cleaved by a member of the ADAM (a disintegrin and metalloproteinase) protease family. A third cleavage, (S3/S4) is mediated by the intramembrane aspartyl protease, γ-secretase. The S3/S4 cleavage liberates the Notch-ICD from the membrane and the ICD is trafficked to the nucleus where it associates with a family of DNA-binding proteins called CSL (CBF1/RBPJ-k/Su (H) Lag) to form a transcriptional complex for activation of specific target genes[8]. In addition to transcellular activation of Notch by Delta ligands, studies have also shown that this ligand can interact with Notch when co-expressed on the same cell and inhibit Notch activity (cis-inhibition)[9–14]. Ubiquitination of DSL ligands including Delta is important for endocytic internalization and recycling to the plasma membrane and may have a role in activation of the ligands prior to Notch receptor binding [3, 15–22].
Endocytic trafficking of DSL ligands is promoted in response to monoubiquitination of the cytoplasmic tail of the ligand by the ubiquitin ligases, Mind bomb (Mib) or Neuralized (Neur) [19, 23–27]. Endocytosis facilitated by Epsin proteins may target the ligand to a specific endocytic compartment that is necessary for DSL ligands to become signaling competent as Epsin mutants move efficiently to the plasma membrane, but do not signal. The exact nature of this activation mechanism is unknown but could involve post-translational modifications to the extracellular domain of the ligand within the compartment or possibly clustering of the ligands to enhance signaling [18, 28–30].
Ubiquitination of proteins is associated with many aspects of cell signaling including protein trafficking, endocytosis, protein interactions, kinase activation and degradation [31–33]. Heuss et al. [18] recently characterized a murine Dll1 in which all 17 lysine (K) residues within the cytoplasmic tail were mutated to arginine (R) (Dll1-K17R) thus rendering the protein resistant to ubiquitinylation. These studies revealed that Dll1-K17R is efficiently endocytosed, but recycling, trans-endocytosis of Notch extracellular domain (Notch-ECD), as well as activation of Notch transcellular signaling are impaired. In addition, Heuss et al. [18] demonstrated wild type Dll1 was partially localized to lipid rafts while Dll1-K17R was excluded from these microdomains, an observation that could account for its failure to activate Notch signaling. Although these studies are elegant and informative, the results from an extensively mutated Dll1 ligand do not address the importance of, or allow identification of specific functions that can be attributed to individual lysine residues. In the current study a panel of mutant Dll1 molecules were examined to investigate the role of specific conserved lysine (K) residues in the cytoplasmic tail of Dll1 and their potential role in Notch signaling, ubiquitination, endocytic recycling and Notch receptor binding. These studies identified a single, conserved lysine (K) residue K613 that is important for both cis- and trans-cellular interactions between Dll1 and Notch to facilitate Notch signaling.
2. Materials and Methods
2.1 Materials
MG132, anti-Vinculin (1:1000; V4505), anti-Flag M2 (1:1000, A2220), anti-Pan Cadherin antibodies (1:1000; C1821/CH-19), protein A beads, protein G beads and protease inhibitor cocktail were purchased from Sigma (St. Louis, MO). Anti-Delta (1:1000; H-265/SC9102), and anti-Notch1 (1:1000; C-20/SC-6014; H-131/sc9170) were from Santa Cruz (Santa Cruz, CA) and have been previously validated [34, 35]. Anti-caveolin-1 (1:1000; 610406) was from BD Biosciences. Anti-β-tubulin (1:500; MAB3408, clone KMX-1) was from Millipore. Absolute QPCR Mixes were from ABgene (Rockford, IL). Fugene 6 transfection reagent was purchased from Roche Diagnostics (Indianapolis, IN). DharmaFect-1 siRNA transfection reagent was from Dharmacon (Lafayette, CO). Notch1/Fc chimera was from R&D (5627-TK) and comprises residues 19–526 from Notch1 fused to the Fc region of human IgG1.
2.2 Plasmids
The construction of full-length p3xFlag-MIB1 has been described previously [36, 37]. Mouse pYX-Asc-Dll1 was purchased from ATCC (Manassas, VA), and pCDNA3-Dll1 was generated by PCR cloning of the coding region of Dll1 into pcDNA3 (Invitrogen). The PCR generated cDNA was sequenced to confirm the fidelity of the reaction and no alterations were identified. The Dll1 lysine mutant constructs were made using site-directed mutagenesis (Stratagene) with mutated oligonucleotide primers corresponding to mutation sites. All mutants were confirmed by sequencing. Vectors for expression of the hemaglutinin-tagged (HA) ubiquitin were kindly provided by Dr. Ted Dawson (Johns Hopkins University). The pGL2-HES1 AB reporter and full length Notch1 constructs were kindly provided by Dr. Nadia Carlesso (Indiana University).
2.3 Cell Culture and Transient Transfection
Wild type mouse embryo fibroblast (MEF) cells were kindly provided by Dr. Maureen A. Harrington (Indiana University School of Medicine, Indianapolis, IN). Human embryonic kidney HEK293 cells (MPBiomedicals) and C3H10T1/2 (10T1/2) cells (ATCC, American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Transient transfection of HEK293 cells was carried out using equal amounts of total plasmid DNA (adjusted with the corresponding empty vectors) and Fugene-6 transfection reagent according to the manufacturer’s guidelines.
2.4 Western Blotting, Immunoprecipitation and Lipid Raft Fractionation
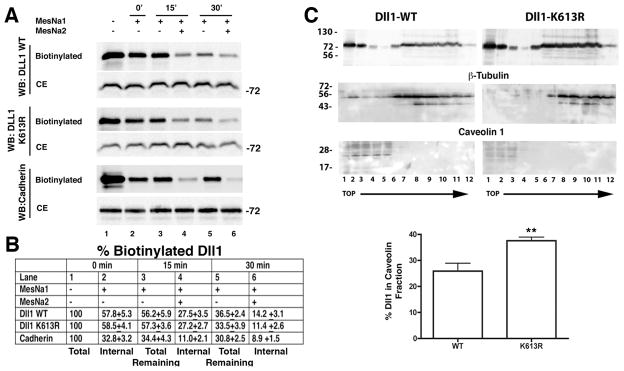
Western blotting and immunoprecipitation were performed as described previously using 7.5% SDS-PAGE [36]. Immunoreactive proteins on Western blots were visualized using the Supersignal West Pico or West Femto detection systems (Pierce) according to manufacturer’s directions. All antibodies were used at a 1:1000 dilution for western blotting except where noted. Cell extracts were prepared in RIPA lysis buffer containing 0.1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 10 mM sodium phosphate, pH 7.2, 2 mM EDTA, 50 mM sodium fluoride, protease inhibitor cocktail and phosphatase inhibitor cocktails. For immunoprecipitation cells were washed with PBS and lysates were prepared in lysis buffer (50 mM Tris-HCl, pH 7.4, 0.1% Nonidet P-40, 1% Triton X-100, 1 mM EGTA, 1 mM EDTA, 0.15 M NaCl) containing a protease inhibitor cocktail. For each immunoprecipitation, clarified cell lysates were incubated with anti-Delta, anti-Flag, anti-HA or IgG (control) antibodies at 4°C for at least 3h. For immunoprecipitations, antibodies were diluted 1:100 in cell lysates containing equivalent amounts of total protein. Protein A or protein G beads were added to the lysates and incubated for further 1.5 h. Immune pellets were washed four times with lysis buffer containing protease inhibitor cocktail, and precipitated proteins were resolved by electrophoresis and then analyzed by Western blotting. Western blotting was quantified using Fuji Imager software (FUJI Medical Systems, USA). Lipid raft fractionation was performed as described by Heuss et al. [18]. HEK293 cells were transfected with plasmids encoding either wild type or K613R mutant Dll1. At 48 h post-transfection, the cells were lysed in 2 mL TN buffer (20 mM Tris, pH7.5; 150 mM NaCl) containing 1% Brij 98 for 30 min on ice. Following passage through a 25-G needle 15 times, the lysates were adjusted to a final concentration of 40% sucrose/1% Brij 98 in TN buffer. Gradients were assembled by the addition of 4 mL of 30% and 4 mL of 5% sucrose in TN buffer. Following centrifugation for 20 h at 230,000 X g, 1 mL fractions were collected from the top to the bottom of the gradient and 40 μL of each fraction was analyzed by western blotting using anti-Delta, anti-β–tubulin and anti-caveolin-1 to identify lipid raft and cytoplasmic fractions.
2.5 Immunoprecipitation of Dll1 from intact cells with N1-Fc
HEK293 cells transiently transfected with vectors for expression of Dll1 were scraped and collected. The cells were washed three times in PBS containing 0.1 mM CaCl2. The washed cells were then incubated in PBS/0.1 mM CaCl2 with 1 μg/mL recombinant Notch1-Fc fusion protein for 18 h at 4°C and then washed three times in PBS/CaCl2 to remove unbound Notch1-Fc before addition of lysis buffer. Equal amounts of lysate were incubated overnight with 50 μL 10% Protein G beads to fractionate the Notch1-Fc/Dll1 complexes.
2.6 In vivo ubiquitination
HEK293 cells were transiently transfected with Dll1 or HA-Ubiquitin constructs as indicated. After 24h, the transfected cells were treated with MG132 for a further 16h. Cell lysates were prepared in RIPA lysis buffer with protease inhibitor cocktail and MG132 (10 μM). Ubiquitinated proteins were immunoprecipitated from the cell extract using anti-HA antibodies and western blotting was performed to detect the presence of Dll1.
2.7 Quantative RT-PCR (qRT-PCR) Analysis of mRNA
RNA was extracted with TRIzol reagent (Invitrogen). 1.2 μg of RNA was used as template for reverse transcription (RT) using Superscript first strand cDNA synthesis kit (Invitrogen) and the resulting cDNAs were dissolved in 20 μL H20. The cDNA levels of specific genes were measured by quantitative real time PCR using Absolute QPCR Mixes (ABgene) and an ABI 7500 Real Time PCR system (Applied Biosystems). The gene- and mouse-specific primers used for QPCR are mHPRT1 5′-GTT ATT GGT GGA GAT GAT CTC TCA ACT-3′ and 5′-TGC AAC CTT AAC CAT TTT GGG GCT G-3′, mHES1 5′-GCT AGA GAA GGC AGA CAT TCT GGA AAT GA-3′ and 5′-CGC GGT ATT TCC CCA ACA CGC T-3′, mHEY1 5′-TCC CTG CTT CTC AAA GGC ACT-3′ and 5′-GGA AAA GAC GGA GAG GCA TCA-3′, mHEY2 5′-AAG CGC CCT TGT GAG GAA A-3′ and 5′-TGT CGG TGA ATT GGA CCT CAT-3′. All samples were amplified in duplicate and every experiment was repeated at least 2 times using biologically independent samples. Relative gene expression was converted using the 2−ΔΔCt method against the internal control HPRT1 housekeeping gene.
2.8 Reporter Gene Assays
Reporter plasmid transfection was carried out using FuGENE6 transfection reagent (Roche) according to the manufacturer’s guidelines. The level of promoter activity was evaluated by measurement of the firefly luciferase activity relative to the internal control TK-Renilla luciferase activity using the Dual Luciferase Assay System essentially as described by the manufacturer (Promega). A minimum of six independent transfections was performed and all assays were performed in duplicate. Results are reported as the mean ± S.E.
2.9 Endocytosis Assay
Endocytosed and exocytosed recycled proteins were identified using a reversible biotinylation assay as previously described[38] with minor modifications (see Supplemental Figure S2 for details).
2.10 Statistical Analysis
Results are expressed as mean ± S.D. The Student’s t test was used to compare quantitative data where indicated. A value of p<0.05 was considered statistically significant.
3. Results
3.1 K613 in the cytoplasmic tail of Dll1 is required for Notch signaling
To identify potential conserved ubiquitination sites within the cytoplasmic tail of Dll1, the sequences of the intracellular domains of DLL from mouse, human and rat were aligned and examined (Supplemental Figure S1). This analysis identified fifteen highly conserved K sites in the cytoplasmic domain of Dll1. A previous study [18] generated a single mutant Dll1 having 17 K residues within the cytoplasmic tail altered to R. Although the complete sequence of the cDNA used in this previous study was not included, the sequence encompassing the specific lysine residues that were mutated was noted. A comparison of the K residues in the previous report and those in our clone and clones from the GenBank database revealed that residue 628 which is K in the Heuss et al [18] Dll1 clone is a highly conserved acidic residue in the mouse clone used for our study as well as rat and human Dll1 clones. In addition, residue 572 is a K residue in both mouse and rat Dll1 clones, but is R in human clones and this residue as well as residue E628 were not targeted for mutation in the mouse Dll1 used in the current study.
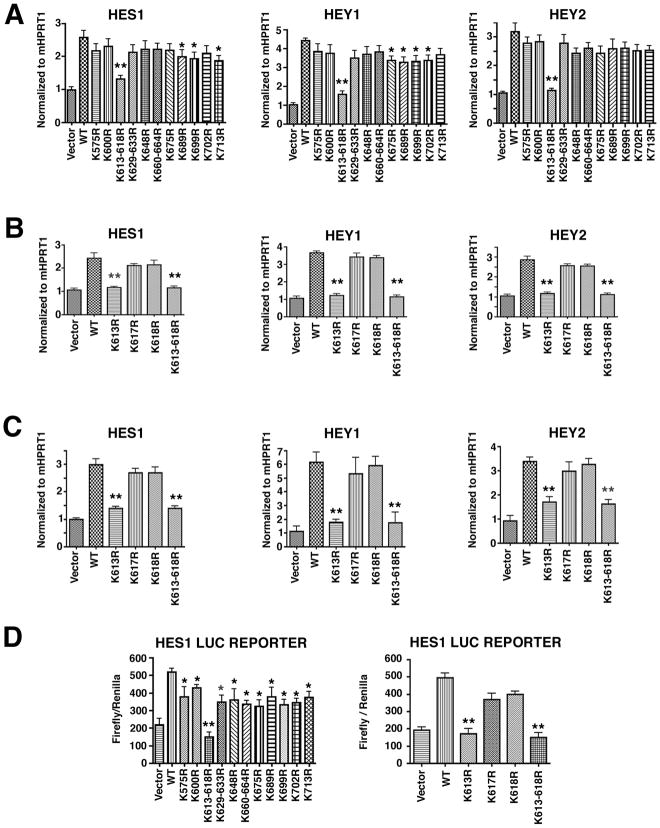
To determine which of these fifteen K residues in the cytoplasmic tail of mouse Dll1 has a role in regulating Notch signaling, a series of Dll1 mutants was generated in which each of these K residues were replaced by arginine (R) either singularly (K575R, K600R, K613R, K617R, K618R, K648R, K675R, K689R, K699R, K702R, K713R) or in combination when conserved K residues were clustered together (K613-618R, K660-664R, K629-633R). The effects of the mutant K to R mouse Dll1 proteins on Notch transcellular signaling were determined using a transcellular co-culture assay. Plasmids encoding murine Dll1 were transfected into human HEK293 cells and after 24h the transfected HEK293 cells were co-cultured with multi-potent mouse 10T1/2 cells or primary mouse embryo fibroblasts, which both express Notch1 receptors on their plasma membranes[39–41]. Using mouse specific primers, qRT-PCR was used to monitor the expression of Notch target genes: HES1, HEY1 and HEY2. Of eleven Dll1 mutants initially tested, only the triple mutant K613R-K618R resulted in a highly significant decrease (p<0.001) in expression of the Notch target genes compared to the wild type Dll1 in 10T1/2 cells (Figure 1A). Expression of three other mutants (Dll1-K689R, -K699R and -K713R) also resulted in attenuated expression of the Notch target genes, although the magnitude of this reduction was smaller and had lower significance (p<0.05). To determine if all three K residues in the triple mutant Dll1-K613-618R were important for expression of Notch target genes, three single mutants were generated using site-directed mutagenesis (Dll1-K613R, -K617R and -K618R). Analysis of these three single mutants using the same HEK293 co-culture approach with 10T1/2 cells (Figure 1B) or with mouse embryo fibroblasts (MEFs) (Figure 1C) revealed only the triple mutant, Dll1-K613-168R and the single mutant, Dll1-K613R were significantly (p<0.001) defective in their ability to activate HES1, HEY1, and HEY2. Finally, activation of a HES1 luciferase reporter plasmid in 10T1/2 cells was significantly attenuated (p<0.001) when co-cultured with HEK293 cells expressing Dll1K613-618R or -K613R as compared to HEK cells expressing wild type Dll1 (Figure 1D). We also observed a small although significant (p<0.05) attenuation in HES1 reporter activity with many of the other K mutant DLL1 molecules (Figure 1D). These data clearly suggest that residue K613 in the cytoplasmic tail of Dll1 is the most important lysine for the transcriptional activation of Notch target genes in a transcellular co-culture assay.
Figure 1. K613 in the cytoplasmic tail of Dll1 is required for Notch signaling.
A–C, HEK293 cells were transfected with wild type Dll1 (WT) or the indicated Dll1 mutants and after 24h, the HEK293 cells were co-cultured with 10T1/2 cells (A–B) or MEF cells (C). After 36h in co-culture, total RNA was prepared and qRT-PCR was carried out to detect the gene expression of mouse HES1, HEY1 and HEY2 using species-specific primers. Samples were amplified in duplicate and each experiment was repeated at least 2–3 times on independent samples, (*p<0.05; **p<0.001) D, Luciferase promoter activation assays were performed using 10T1/2 cells that had been transfected with a luciferase HES1 reporter plasmid for 12h before co-culture with HEK293 cells transfected with wild type or the indicated Dll1 mutants. Cell lysates were prepared and luciferase activity was measured as described in methods. Luciferase values are presented as relative luciferase activity compared to empty expression vector which was normalized to 1 and are the mean ± S.E. of 6 samples, *p<0.05; **p<0.001. Statistical significance was determined by the Student’s t test.
3.2 Ubiquitination of Dll1-K613R is altered
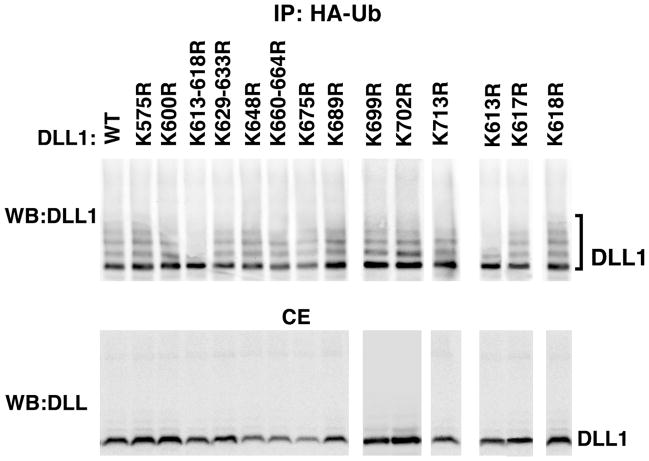
As the results in Figure 1 suggested that Dll1-K613 was important for transcriptional activation of several Notch1 genes, the ubiquitination profile of the Dll1-K613-617R and Dll1-K613R mutants was compared to wild type Dll1 as well as the remaining panel of mutant Dll1 molecules (Figure 2). Lysates from HEK293 cells transfected with vectors for expression of either with wild type or mutant Dll1, HA-ubiquitin, and Mib1 were examined by western blotting to detect Dll1 in total ubiquitinated protein fractions. This analysis revealed that the ubiquitination profile of Dll1-K613-617R and Dll1-K613R was altered compared to the wild type Dll1 and the remaining Dll1 mutants. Specifically it was noted that only a single mono-ubiquitinated species was detected and the typical ubiquitin ladder that is found in the remaining Dll1 mutants was strongly attenuated.
Figure 2. Ubiquitination of Dll1 K613 is altered.
HEK293 cells were transfected with HA-ubiquitin, Mib1, and wild type or mutant Dll1 as indicated. At 24 h the cells were treated with 10 μM MG132 for an additional 16 h to allow accumulation of ubiquitinated proteins before lysis. HA-tagged ubiquitinated proteins were immunoprecipitated using anti-HA antibody and Dll1 was detected by western blotting using anti-Delta antibody (sc-9102). To confirm the presence of the relevant proteins, an aliquot representing 1/20th of the total cell extract (CE) was reserved prior to immunoprecipitation and analyzed in parallel for expression of Dll1 (lower panel). A representative blot is shown, n=3. IP, immunoprecipitation; WB, western blotting; Ub, ubiquitin; WT, wild type.
3.3 Dll1-K613R interacts with Mib1
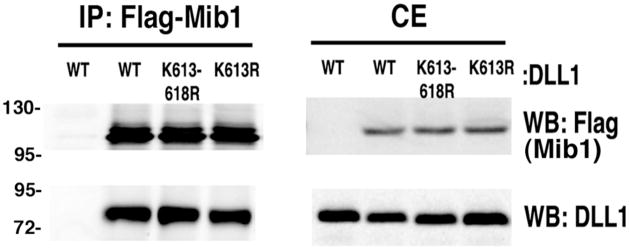
To determine whether or not the attenuated ubiquitination of Dll1-K613-618R or K613R resulted from altered binding between the ubiquitin ligase Mib1 and the mutant Dll1 proteins, HEK293 cells were transfected with plasmids for expression of flag tagged Mib1 and Dll1 wild type or mutants as indicated (Figure 3). After 24h, the cells were lysed and Mib1 was immunoprecipitated using anti-Flag antibody. The immune complexes were then analyzed by western blotting to determine if Dll1 was present (Figure 3). The results of this co-immunoprecipitation experiment demonstrated that both Dll1-K613-618R and Dll1-K613R mutants interact with Mib1 similar to wild type Dll1.
Figure 3. Dll1-K613R interacts with Mib1.
HEK293 cells were co-transfected with Mib1 and either DLL1-WT, DLL1-K613-618R, or DLL1-K613R plasmids as indicated. After 36 h, Mib1 was immunoprecipitated using anti-Flag antibody. Western blots were probed with anti-Flag or anti-Delta (sc-9102) antibodies to detect Mib1 or DLL1. Shown is a representative blot, n=3. IP, immunoprecipitation; WB, western blotting; CE, cell extract; WT, wild type.
3.4 Residue K613 in Dll1 is important for interaction with Notch1 Extracellular Domain
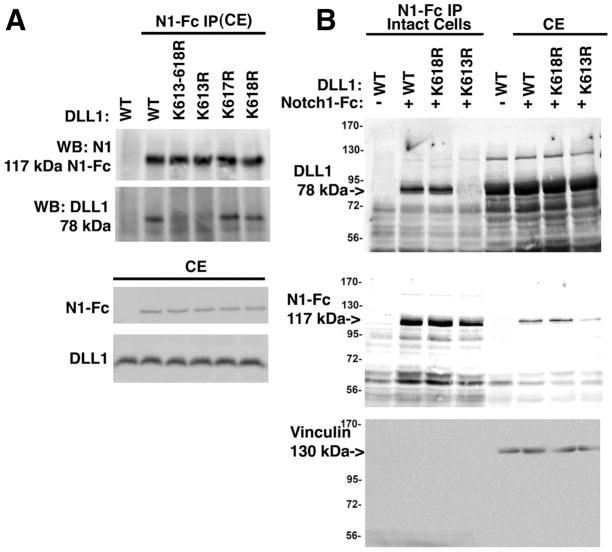
A key step in the activation of Notch target genes and inhibition of Notch signaling is physical interaction between Dll1 ligands and Notch receptors both in trans and cis [9–12, 14, 19, 42]. To determine if the binding between Notch1 and Dll1 or Dll1-K613R was altered, the association of these molecules was examined using purified, recombinant Notch1/Fc to immunoprecipitate Dll1 both in cis (from lysed cells) and trans (from intact cells) configurations. Notch1/Fc is a chimeric fusion protein comprised of Notch1-ECD (residues 19-526) fused to Fc region of human IgG and can be used as a source of Notch1-ECD. To determine if cis binding between Notch and Dll1 is altered, HEK293 cells transiently expressing either wild type or mutant Dll1 were lysed and the cell lysate was then incubated with purified, recombinant Notch1/Fc (Figure 4A). Analysis of Notch1/Fc immune complexes by western blotting revealed that only wild type Dll1 or the mutants K617R and K618R Dll1 bound Notch1/Fc while K613-618R or K613R did not (Figure 4A). These data suggest that cis interactions between Notch and the mutant Dll1-K613-618R or Dll1-K613R are significantly impaired compared to wild type Dll1. To determine if trans binding between Notch and Dll1 is also altered, Notch1/Fc was incubated with intact cells and then the remaining unbound Notch1/Fc was washed before cell lysis and addition of protein G (Figure 4B). Dll1-K613R was not present in Notch1/Fc immune complexes from intact cells (Figure 4B) suggesting that trans interactions between Notch and the Dll1-K613R mutant are attenuated. Western blotting of input cell extracts confirmed the presence of Dll1 and Notch1/Fc in cell lysates. These results demonstrate the importance of K613 in Dll1 for binding to the Notch1-ECD fusion protein both in whole cell lysates (cis) and to Dll1 on the surface of intact cells (trans).
Figure 4. Residue K613 in Dll1 is important for both cis and trans interactions with Notch1.
A, HEK293 cells were transfected with wild type or mutant Dll1 as indicated. After 36 hrs. cell lysates were prepared and then incubated with 1 μg/mL recombinant Notch1-Fc fusion protein pre-complexed to protein G agarose. Western blotting was carried out to detect Dll1 and Notch1-Fc using anti-Delta (sc-9102) or anti-Notch1 (sc-9170) antibodies. To confirm the expression of proteins western blotting of 25 μg total cell extract was used. B, HEK293 cells were transfected with wild type or mutant Dll1 as indicated. After 36 h, the intact cells were incubated with 1 μg/mL recombinant Notch1-Fc fusion protein for 18h at 4°C, the intact cells were washed to remove unbound Notch1-Fc, lysed and then equal amounts of total protein were incubated with protein G agarose. To confirm the presence of the relevant proteins, an aliquot representing 25 μg of the total cell extract (CE) was reserved prior to immunoprecipitation and analyzed in parallel. Western blotting was carried out as in ‘A’ and vinculin was used to confirm that equivalent amounts of CE were used for analysis. Representative blots are shown for each panel; n=3. IP, immunoprecipitation, CE, cell extract, N1-Fc, Notch1 extracellular domain-Fc fusion protein.
3.5 Mutation of Dll1 K613 does not alter its membrane endocytosis or recycling, but does alter lipid raft localization
Previously, a Dll1 mutant that was unable to be ubiquitinylated was shown to be trafficked to the plasma membrane efficiently, but to be defective in membrane recycling and excluded from lipid raft microdomains[18]. Based on this, we sought to determine if Dll1-K613R alters trafficking to and from the plasma membrane using an assay to measure the apparent rate of trafficking of the wild type (WT) and mutant Dll1-K613R to the plasma membrane [38]. In this assay the relative changes in the internalized, biotin-labeled pool of Dll1 following incubation periods at either 4°C (no recycling) or 37°C (recycling) were determined (see Materials and Methods and Supplemental Figure S2A for details). Preliminary experiments using intact HEK293 cells established that within 30 min at 37°C, biotinylation of Dll1 is maximal and that MesNa treatment was able to effectively strip biotin from labeled surface proteins (Supplemental Figure S2B, C). To determine the changes in the relative level of the internal, biotinylated pools of Dll1 as an indicator of the rate of endocytosis, wild type Dll1-WT or mutant Dll1-K613R were expressed in HEK293 cells and the surface membrane proteins of intact cells were biotinylated for 30 min at 37°C to allow for cycling and maximal labeling of the various pools of membrane proteins. Biotinylated proteins were purified using streptavidin conjugated beads and analyzed by western blotting. For all the blots in Figure 5A, the intensity of the Dll1 bands in lanes 2–6 was first normalized to the total Dll1 shown in the CE blot and then compared to the biotinylated Dll1 present in the streptavidin fraction following the labeling period (Figure 5A, lane 1). To determine the fraction of the total biotinylated pool that was located in an internal compartment, the labeled cells were incubated at 4°C to block further endocytic recycling and MesNa was used to strip the biotin from the exposed plasma membrane proteins (MesNa1) before cell lysis. This leaves only the internalized Dll1 labeled with biotin (Figure 5A, lane 2). Cells were then returned to 37°C for either 15 or 30 min to allow the internalized biotinylated Dll1 to recycle back to the plasma membrane or to be degraded (Figure 5A, lanes 3,5). In parallel, cells were subjected to a second round of stripping (MesNa2) following this 37°C incubation to remove the labeled Dll1 that had recycled to the plasma membrane (Figure 5A, lanes 4,6). The decreased Dll1 signal seen between lanes 3 and 4 and between lanes 5 and 6 thus represents that amount of biotinylated Dll1 that had recycled back to the plasma membrane during the 15 or 30 minute incubation at 37°C, respectively. The signals on the western blots (obtained from 3 separate experiments) were quantitated by densitometry and compared to the total biotinylated Dll1 (lane 1) and the percent Dll1 remaining in the biotinylated fraction is shown in the table (Figure 5B).
Figure 5. Mutation of Dll1 K613 does not alter membrane endocytosis or recycling, but does alter lipid raft localization.
A, HEK293 cells were transfected with plasmids for expression of wild type (WT) or mutant Dll1-K613R as indicated, and the trafficking of Dll1 between internal compartments and the plasma membrane was determined as described in “Materials and Methods” and supplementary figure S2. The biotinylated Dll1 in streptavidin fractions was detected using anti-Delta antibodies (sc-9102). Cadherins are known to undergo endocytosis and recycling and serve as a control. Cadherins were detected using an anti-Pan-cadherin antibody (CH-19). Shown are representative blots, n=3. CE, cell extract, represents 1/20th of the total cell extract prior to streptavidin fractionation; WB, western blot. B, Data shown in ‘A’ were quantitated from 3 separate experiments, and the percent ± SD (%) of biotinylated Dll1 present in each group shown. These values were determined by first normalizing the biotinylated Dll1 in the streptavidin fraction to the amount of Dll1 in the corresponding CE lane and then comparing this value to total biotinylated Dll1 (lane 1). C, Western blotting of HEK293 cells transfected with either wild type (WT) or K613R mutant Dll1. At 40 post-transfection, cells were mechanically lysed in buffer containing 1% Brij98 and then fractionated by ultracentrifugation through a sucrose step-gradient as described in “Materials and Methods”. Fractions were collected from top (fraction 1) to the bottom (fraction 12) of the gradient. Antibodies to β-tubulin (55 kDa), or caveolin-1 (24 kDa) were used to define cytoplasmic (detergent soluble) or lipid raft (detergent insoluble) fractions respectively, in the gradient. Bands were quantitated by densitometry and the percentage of Dll1 present in the caveolin positive fractions determined. Mean data ± SD obtained from 3–4 independent experiments are shown in the graph. **indicates statistical significance; p<0.05, Students t test.
Approximately 57.8±5.3 and 58.5±4.1% of the pool of biotinylated wild type or mutant Dll1 respectively was sequestered in an internal compartment following labeling (0 min at 37°C). The amount of the internalized biotinylated Dll1 that recycled back to the plasma membrane after 15 or 30 minute incubation at 37°C was similar for both wild type and K613R mutant Dll1. At 15 minutes 27.5±3.5% of the wild type and 27.2±2.7% of the mutant remained in the internal compartment (Figure 5A) and after 30 minutes 14.2±3.1% of the wild type and 11.4±2.6% of the mutant remained in the internal compartment. During this recycling period it was also noted that there was a significant decrease in the total biotinylated wild type (57.8 vs. 36.5%) and mutant Dll1 (58.5 vs. 33.5%) following 30 minutes of incubation at 37°C (Figure 5A,B lane 2 and lane 5). This decrease likely represents degradation of Dll1 during the experiment. The decrease determined for both wild type and K613R is not statistically significant (36.5±2.4 vs. 33±3.9%; Figure 5A,B, lane 5), suggesting that K613R is not degraded more rapidly than wild type Dll1. In control experiments, the levels of the internal pool of biotinylated cadherins detected by a pan-cadherin antibody also remained relatively stable during this period (32.8±3.2 vs. 30.8±2.5%; Figure 5A,B, lanes 2 and 5). Together, these results suggest that mutation of K613 in Dll1 does not significantly alter the rate of its cycling between the plasma membrane and internal compartments or its stability compared to wild type Dll1.
To determine if mutation of residue K613 alters lipid raft microdomain localization, the fraction of Dll1 WT or K613R localized to caveolin-1 containing membrane fractions was determined (Figure 5C). Membranes were isolated from HEK cells transfected with wild type or K613R mutant Dll1 and fractionated on sucrose density gradients [18]. Membrane fractions were analyzed by western blotting to determine how much Dll1 was present in the caveolin-1 rich lipid raft fractions. As expected, caveolin (24 kDa) was restricted to fractions located at the top of the gradient. There was some small experimental variation in the number of fractions in which caveolin was readily detectable ranging from fractions 1–3 to fractions 1–5, likely reflecting small differences in the preparation and collection of the gradient and the total amount of protein loaded. Densitometric analysis comparing the amounts of Dll1 WT or K613R in caveolin-1 containing fractions revealed that there was a significant difference in their distribution. This analysis showed that 25.9 ± 3.0% of Dll1 WT compared to 37.5 ± 2.6% of K613R is in caveolin-1 containing fractions. These results suggest that Dll1-K613R is more efficiently recruited to lipid rafts or alternatively, is less effectively endocytosed from lipid rafts.
4.0 Discussion
These studies demonstrate that residue K613 in the cytoplasmic tail of Dll1 is critically important for activation of Notch signaling. Mutation of Dll1-K613 to R attenuated transcellular Notch1 signaling and activation of target genes in co-cultured cells. The ubiquitin profile of this mutant is altered such that multi-ubiquitination is greatly attenuated, despite the fact that Dll1-K613R interacts with at least one E3 ubiquitin ligase, Mib1. Dll1-K613R is also unable to physically interact with Notch1 receptors either in cis or trans configurations, although it is expressed at the cell surface, endocytosed and degraded at a similar rate to wild type Dll1. Interestingly, there is a higher level of Dll1-K613R present in lipid raft microdomains, suggesting that this mutation may alter endocytosis from rafts or alternatively enhance association with these microdomains.
Several possible mechanisms could explain how mutation of K613 within the cytoplasmic tail of Dll1 profoundly reduces the activation of Notch dependent genes. These include alteration to ubiquitination, trafficking to and from the membrane, inability to interact with Notch receptors or ubiquitin ligases, and stability or degradation. Examination of each of these possibilities led to the discovery that Dll1-K613R has an altered pattern of ubiquitination and an impaired ability to interact with Notch receptors (Figures 2 and 4). In contrast, Dll1-K613R interacted with at least one ubiquitin ligase Mib1, (Figure 3), and was localized to the plasma membrane and underwent membrane recycling (Figure 5) similar to wild type Dll1.
The finding that global recycling and stability of Dll1-K613R is not significantly different to wild type led to the examination of the fraction of this mutant present in lipid raft microdomains. Interestingly, the fraction of Dll1-K613R present in these microdomains is greater than wild type Dll1. This result can be interpreted in at least two ways, which are not mutually exclusive. First, it is possible that recruitment of Dll1-K613R to lipid microdomain is enhanced because of structural differences related to either the residue change or altered ubiquitination. If indeed structural alterations are responsible, they are likely subtle as the degradation of this mutant is similar to wild type Dll1. Alternatively, it is possible that endocytosis from lipid rafts is attenuated, even though global endocytosis of Dll1-K613R is not significantly different from wild type Dll1. This may suggest that recycling from lipid microdomains is a key event in “activation” of Dll1 ligand to a state competent to interact with Notch receptor. Although the steps leading to “activation” of DSL ligands are poorly understood, endocytosis is thought to be a important for generation of an “active” ligand that is competent for a productive interaction with Notch receptors to initiate signaling [18, 43, 44]. The current studies further refine this paradigm, suggesting the possibility that endocytic recycling from lipid microdomains is a key event to ligand activation.
Comparison of the ubiquitination pattern of wild type Dll1 to the panel of Dll1 mutants revealed that they all appear to be mono-ubiquitinated, as evidenced by the prominent ubiquitin band detected by anti-Dll1 antibody in the ubiquitinated fraction of cellular proteins. At least 2–3 additional ubiquitinated forms are also consistently evident in this panel of mutants resembling a ladder of multi-ubiquitinated Dll1 species. The only exception to this ubiquitination pattern in this panel of conserved K mutants is Dll1-K613, which lacks this laddering pattern, although it does appear to be mono-ubiquitinated. This finding raises the possibility that loss of one ubiquitination site promotes ubiquitination of one of the other K residue(s) in the cytoplasmic tail of Dll1 and could account for the failure to identify a dominant ubiquitination site. It is also possible that K572 in mouse and rat Dll1, which is not conserved in human sequences and therefore was not analyzed in our study, could be an important murine specific ubiquitination site. However, as there are no other non-conserved K residues in the human Dll1 cytoplasmic tail that could correspond to mouse K572, it is difficult to see how the additional K at position 572 in mice and rats can be of major physiological importance.
The findings of the current study, complement and extend those of Heuss et al [18]. In this previous study, all of the K residues in the intracellular domain of a mouse Dll1 were mutated to R to generate a “ubiquitin-less” mutant (K17R). The K17R mutant exhibited a complete lack of ubiquitination, did not activate Notch signaling, and although this mutant was endocytosed, its endocytic recycling back to the plasma membrane was severely attenuated. In contrast to the current results, the ubiquitinless K17R mutant was completely excluded from lipid raft microdomains. These previous results suggested the possibility that ligand recycling and lipid raft localization of Dll1 are both required for activation of Notch signaling[19]. The current studies demonstrate that some of the defects observed in the K17R mutant Dll1 are separable and can be attributed to functions of different lysine residues within the Dll1 cytoplasmic tail. For example, K613 is critical for Notch signaling, and its mutation results in a loss of the observed multi-ubiquitination of Dll1, but does not alter the endocytic recycling of Dll1-K613R to the plasma membrane or its stability and increases its association with lipid raft microdomains. In addition, it appears that endocytic recycling and notch binding can be separated based on the finding that Dll1-K613R is recycled as efficiently as wild type Dll1 but is unable to interact with and signal to Notch receptors.
5.0 Conclusion
Collectively, the results of the current studies demonstrate that residue K613 on the cytoplasmic tail of Dll1 is required for both cis and transcellular interactions between Dll1 and Notch1 receptors, as well as for activation of Notch signaling. Although the endocytic recycling and stability of Dll1-K613R is not altered, its localization to lipid raft microdomains is enhanced. Dll1-K613R binds Mib1 and although it is still mono-ubiquitinated, the typical multi-ubiquitination pattern is reduced. These studies identify K613 of Dll1 as a key focal point for regulating Notch signaling and endocytic activation of Notch ligands.
Supplementary Material
Research Highlights.
Seventeen conserved lysine residues in the cytoplasmic tail of Delta like1 (Dll1) were identified and mutated.
Notch signaling, ubiquitination, endocytic recycling, lipid raft localization and Notch binding functions were examined.
K613 was identified as important for Notch signaling.
K613R altered the multiubiquitination profile of Dll1.
K613R did not alter endocytosis or stability of Dll1 but did alter binding to Notch1 and lipid raft localization.
Acknowledgments
This work was supported by grants from the National Institutes of Health HL54118 & DK062810 to P. J. G. and DK63110 to B.P.H. The IUSM Cancer Biology Training Program T32-CA111198 and a DeVault Fellowship supported R.C.W
Abbreviations
- Dll1
Delta-like 1
- DSL
Delta/Serrate/Lag2
- Notch-ICD
Notch Intracellular domain
- Notch-ECD
Notch extracellular domain
- HES1
Hairy/enhancer of Split 1
- HEY1
Hairy/enhancer of split related with YRPW motif protein 1
- CSL
Suppressor of Hairless/Lag1
- siRNA
silencing RNA
- IP
immunoprecipitation
- WB
western blot
- HA
hemaglutinin
- MesNa
2-mercaptoethanesulfonic acid sodium salt
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 2.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 3.D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raymond T, Schaller M, Hogaboam CM, Lukacs NW, Rochford R, Kunkel SL. Toll-like receptors, Notch ligands, and cytokines drive the chronicity of lung inflammation. Proc Am Thorac Soc. 2007;4:635–641. doi: 10.1513/pats.200706-067TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush G, diSibio G, Miyamoto A, Denault JB, Leduc R, Weinmaster G. Ligand-induced signaling in the absence of furin processing of Notch1. Dev Biol. 2001;229:494–502. doi: 10.1006/dbio.2000.9992. [DOI] [PubMed] [Google Scholar]
- 6.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, Aster JC. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopan R. Notch: a membrane-bound transcription factor. J Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- 9.de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- 10.Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 11.Klein T, Brennan K, Arias AM. An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev Biol. 1997;189:123–134. doi: 10.1006/dbio.1997.8564. [DOI] [PubMed] [Google Scholar]
- 12.Miller AC, Lyons EL, Herman TG. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr Biol. 2009;19:1378–1383. doi: 10.1016/j.cub.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, Redfield C, Baron M, Lea SM, Handford PA. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Casey Corliss D. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev Biol. 2004;267:361–373. doi: 10.1016/j.ydbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, Li Q, Jiang YJ. Zebrafish Mib and Mib2 are mutual E3 ubiquitin ligases with common and specific delta substrates. J Mol Biol. 2007;366:1115–1128. doi: 10.1016/j.jmb.2006.11.096. [DOI] [PubMed] [Google Scholar]
- 18.Heuss SF, Ndiaye-Lobry D, Six EM, Israel A, Logeat F. The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc Natl Acad Sci U S A. 2008;105:11212–11217. doi: 10.1073/pnas.0800695105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- 20.Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 2005;3:e96. doi: 10.1371/journal.pbio.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai EC. Protein degradation: four E3s for the notch pathway. Curr Biol. 2002;12:R74–78. doi: 10.1016/s0960-9822(01)00679-0. [DOI] [PubMed] [Google Scholar]
- 22.Kanwar R, Fortini ME. Notch signaling: a different sort makes the cut. Curr Biol. 2004;14:R1043–1045. doi: 10.1016/j.cub.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Struhl G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 2005;132:2883–2894. doi: 10.1242/dev.01860. [DOI] [PubMed] [Google Scholar]
- 24.Deblandre GA, Lai EC, Kintner C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev Cell. 2001;1:795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- 25.Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- 26.Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. neuralized Encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev Cell. 2001;1:807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- 27.Yeh E, Dermer M, Commisso C, Zhou L, McGlade CJ, Boulianne GL. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr Biol. 2001;11:1675–1679. doi: 10.1016/s0960-9822(01)00527-9. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- 29.Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. The Journal of cell biology. 2009;185:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratt EB, Wentzell JS, Maxson JE, Courter L, Hazelett D, Christian JL. The cell giveth and the cell taketh away: an overview of Notch pathway activation by endocytic trafficking of ligands and receptors. Acta histochemica. 2011;113:248–255. doi: 10.1016/j.acthis.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varshavsky A. Regulated protein degradation. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 33.Malynn BA, Ma A. Ubiquitin makes its mark on immune regulation. Immunity. 2010;33:843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 35.Dyczynska E, Sun D, Yi H, Sehara-Fujisawa A, Blobel CP, Zolkiewska A. Proteolytic processing of delta-like 1 by ADAM proteases. J Biol Chem. 2007;282:436–444. doi: 10.1074/jbc.M605451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Y, Blue EK, Dixon S, Hou L, Wysolmerski RB, Gallagher PJ. Identification of a new form of death-associated protein kinase that promotes cell survival. J Biol Chem. 2001;276:39667–39678. doi: 10.1074/jbc.M101886200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin Y, Blue EK, Dixon S, Shao Z, Gallagher PJ. A death-associated protein kinase (DAPK)-interacting protein, DIP-1, is an E3 ubiquitin ligase that promotes tumor necrosis factor-induced apoptosis and regulates the cellular levels of DAPK. J Biol Chem. 2002;277:46980–46986. doi: 10.1074/jbc.M208585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fournier KM, Gonzalez MI, Robinson MB. Rapid trafficking of the neuronal glutamate transporter, EAAC1: evidence for distinct trafficking pathways differentially regulated by protein kinase C and platelet-derived growth factor. J Biol Chem. 2004;279:34505–34513. doi: 10.1074/jbc.M404032200. [DOI] [PubMed] [Google Scholar]
- 39.Zhao L, Li G, Chan KM, Wang Y, Tang PF. Comparison of multipotent differentiation potentials of murine primary bone marrow stromal cells and mesenchymal stem cell line C3H10T1/2. Calcif Tissue Int. 2009;84:56–64. doi: 10.1007/s00223-008-9189-3. [DOI] [PubMed] [Google Scholar]
- 40.Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2001;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, Kensler TW. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordle J, Redfieldz C, Stacey M, van der Merwe PA, Willis AC, Champion BR, Hambleton S, Handford PA. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J Biol Chem. 2008;283:11785–11793. doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- 43.Le Borgne R, Schweisguth F. Notch signaling: endocytosis makes delta signal better. Curr Biol. 2003;13:R273–275. doi: 10.1016/s0960-9822(03)00199-4. [DOI] [PubMed] [Google Scholar]
- 44.Le Borgne R, Schweisguth F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev Cell. 2003;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.