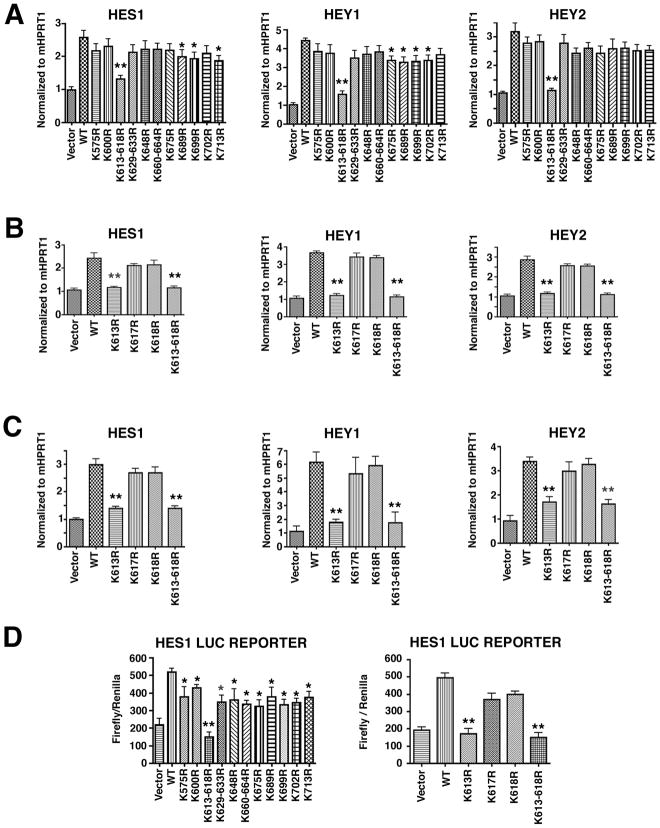
Figure 1. K613 in the cytoplasmic tail of Dll1 is required for Notch signaling.
A–C, HEK293 cells were transfected with wild type Dll1 (WT) or the indicated Dll1 mutants and after 24h, the HEK293 cells were co-cultured with 10T1/2 cells (A–B) or MEF cells (C). After 36h in co-culture, total RNA was prepared and qRT-PCR was carried out to detect the gene expression of mouse HES1, HEY1 and HEY2 using species-specific primers. Samples were amplified in duplicate and each experiment was repeated at least 2–3 times on independent samples, (*p<0.05; **p<0.001) D, Luciferase promoter activation assays were performed using 10T1/2 cells that had been transfected with a luciferase HES1 reporter plasmid for 12h before co-culture with HEK293 cells transfected with wild type or the indicated Dll1 mutants. Cell lysates were prepared and luciferase activity was measured as described in methods. Luciferase values are presented as relative luciferase activity compared to empty expression vector which was normalized to 1 and are the mean ± S.E. of 6 samples, *p<0.05; **p<0.001. Statistical significance was determined by the Student’s t test.