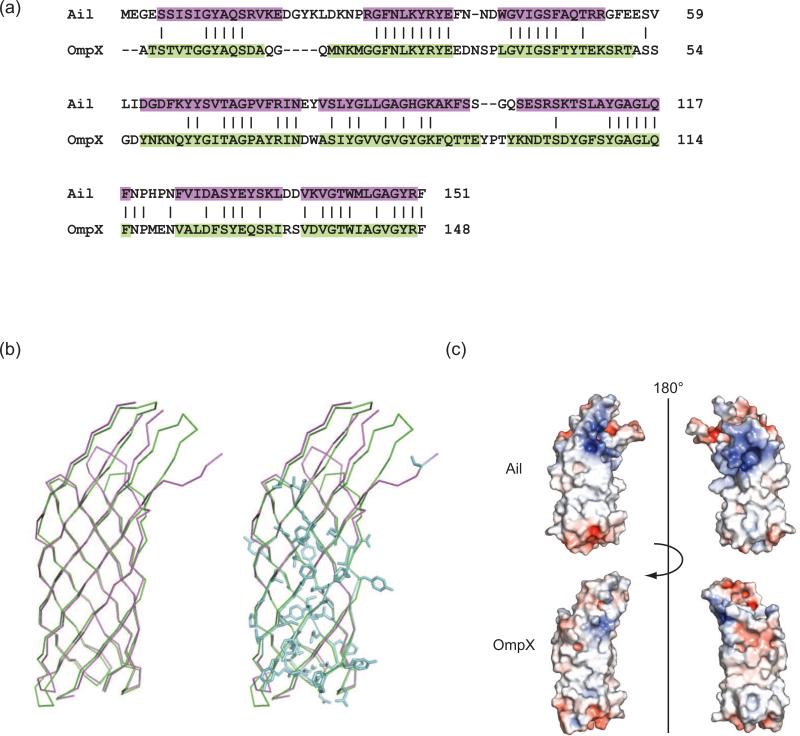
Figure 3. Structure-based sequence alignment of Ail and OmpX and electrostatic surface properties.
(a) Dalilite (Baker et al, 2001) structure-based sequence alignment of Ail (purple) with E. coli OmpX (green) (PDB ID = 1QJ8). (b) Superposition of Ail (purple) with OmpX (green) using the alignment in (a) identified 65 identical residues (blue) shared between the two proteins for an overall sequence identity of 41%. Nearly all of these conserved residues are located in the transmembrane β-strands of the barrel. Since Ail residues involved in adhesion and complement resistance are located primarily in the extracellular loops, this may explain why OmpX does not have similar functions. (c) Electrostatic surface representations for Ail and OmpX. Regions with potentials above +5kT (blue) and below -5kT (red) were calculated by the program APBS (Holm & Park, 2000). Ail displays two regions of positive charge (blue) on the extracellular surface, approximately separated by a 180° rotation. In contrast, the surface of OmpX displays relatively little positive or negative charge.