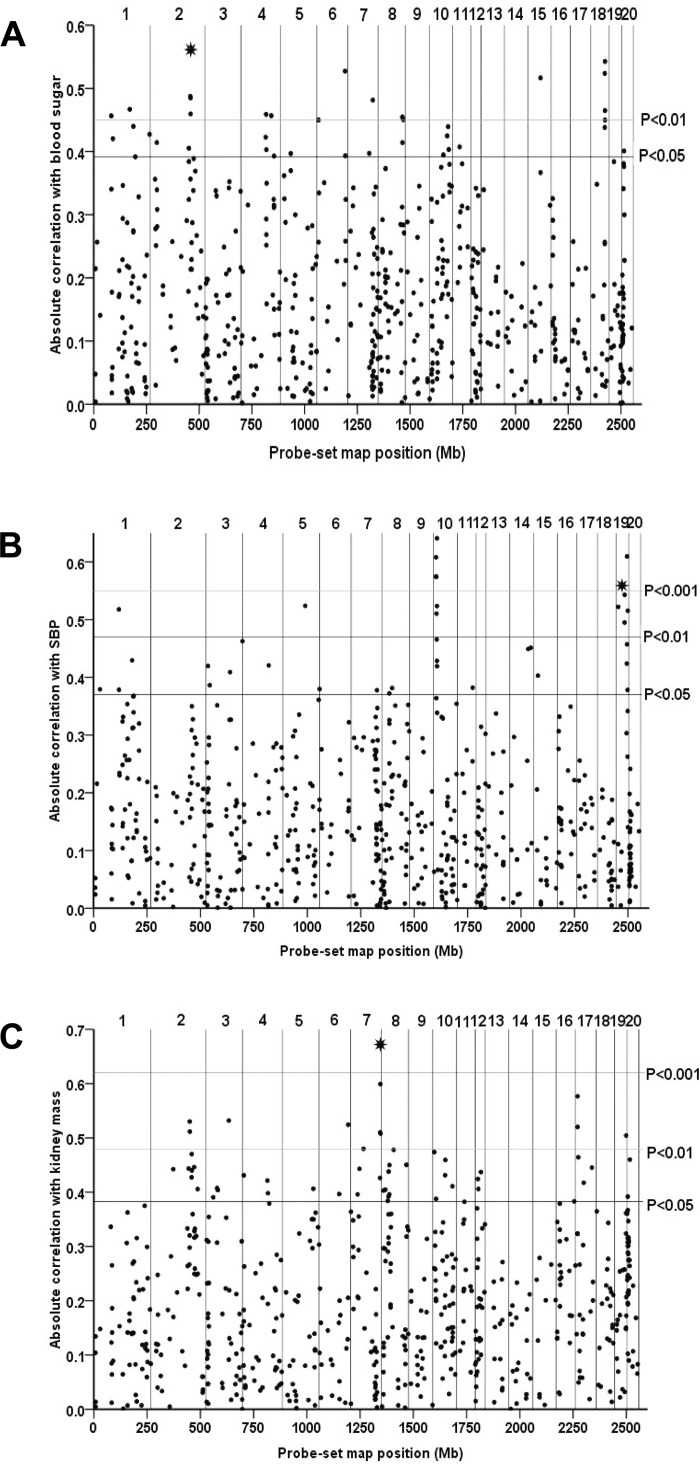
Fig. 3.
Examples of quantitative trait transcript analysis of cis-eQTL genes and physiological and metabolic traits in the rat. Expression profiles of cis-eQTL genes (genome-wide P value ≤ 0.05) across the BXH/HXB recombinant inbred (RI) strains were correlated with values for blood sugar (oral glucose tolerance test, 120 min after glucose) (adrenal cis-eQTL dataset) (A), systolic blood pressure (adrenal cis-eQTL dataset) (SBP) (B), and kidney mass (fat cis-eQTL dataset) (C). For each cis-eQTL, the absolute Pearson correlation coefficient between expression values and phenotype was plotted against the location of the probe set (Mb). Vertical lines mark the physical position of the end of each chromosome. The number of each chromosome is shown above each plot. Horizontal lines indicate empirical significance levels P = 0.05, P = 0.01, and P = 0.001 of the correlations. Prioritized, qPCR validated cis-eQTL genes with transcripts showing strong Pearson correlation coefficients for the respective traits are shown by an asterisk [A: Pex11b (chromosome 2), B: Hp (chromosome 19), C: Cela1 (chromosome 7)].