Abstract
The delivery of nucleic acids has the potential to revolutionize medicine by allowing previously untreatable diseases to be clinically addressed. Viral delivery systems have shown immunogenicity and toxicity dangers, but synthetic vectors have lagged in transfection efficiency. Previously, we have developed a modular, linear-dendritic block copolymer architecture with high gene transfection efficiency compared to commercial standards. This rationally designed system makes use of a cationic dendritic block to condense the anionic DNA and forms complexes with favorable endosomal escape properties. The linear block provides biocompatibility, protection from serum proteins, and can be functionalized with a targeting ligand. In this work, we quantitate performance of this system with respect to intracellular barriers to gene delivery using both high-throughput and traditional approaches. An image-based, high throughput assay for endosomal escape is described and applied to the block copolymer system. Nuclear entry is demonstrated to be the most significant barrier to more efficient delivery and will be addressed in future versions of the system.
Introduction
Nucleic acid therapies have unique potential as a transformative element in clinical medicine over the coming decades. The primary barrier to clinical application of gene therapy has been the lack of safe, efficient materials to deliver genes to appropriate tissues (1, 2). A variety of viral vectors, including adenoviruses, adeno-associated viruses (AAVs), and retroviruses have been studied as gene delivery agents (3). While these materials can achieve high levels of protein expression, they have suffered from concerns of safety, immunogenicity, scale-up for manufacturing, and limited size of the delivered gene (1). When viral vectors are used for transient gene expression, the increased immune response upon repeat injections becomes a limiting factor (4, 5).
Synthetic systems have been developed as a safer alternative to viral transfection, but have struggled to achieve efficiency (protein produced per plasmid) on the same order of magnitude as viral vectors. Synthetic delivery systems include cationic polymers (6, 7), liposomes (8–10), and dendrimers (11, 12), among other material classes. Polymeric systems developed include linear and branched polyethyleneimine (PEI) (13), linear and dendritic polyamidoamine (PAMAM) (11), poly-β-amino esters (PBAE) (14–16), as well as many other linear and branched polyamines (17). The molecular design of such polymeric systems must overcome several extracellular and intracellular barriers to achieve therapeutic levels of protein expression. In the bloodstream, DNA complexes must avoid hepatic and renal clearance, detection and removal by the immune system, binding to charged serum proteins, inter-complex aggregation, degradation by plasma nucleases and uptake by non-targeted somatic cells (18–21). Complexes then must be taken up efficiently by the cells of interest, and escape the endosomal compartment into which they are initially trafficked. Failure to do so results in degradation in the lysosome, recycling to the cell membrane, and lack of transfection. Once out of the endosome, the complex must translocate to the nucleus and cross the nuclear membrane. Unpackaging of the DNA from the complex is also necessary prior to expression of the delivered plasmid by the host cell. As synthetic vectors do not contain transcription factors as viruses do, the final expression of uncomplexed plasmids in the nucleus has been shown to be a bottleneck as well (24).
In order to address these barriers to transfection, our lab has previously developed a linear-dendritic block-copolymer system, in which a generation 5.0 PAMAM dendron is conjugated to a polyethylene glycol (PEG) linear block, which itself is end-linked to a targeting moiety (25, 26). This PAMAM-G5-PEG system performed well in vitro, with transfection efficiencies nearly ten-fold greater than commercially available branched PEI, and it displayed targeted, receptor-mediated uptake with cytotoxicities significantly lower than unmodified PAMAM or PEI. The targeting moiety on the targeted PAMAM-G5-PEG conjugates is a short peptide, WIFPWIQL, identified from in vivo phage display techniques by the Arap/Pasqualini lab (27). WIFPWIQL binds GRP78/BiP/HSP70 – a glucose-response protein found intracellularly at the endoplasmic reticulum (ER) in benign cell types, but on the cell membrane of many solid tumor cell types (28, 29). In our previous publication, we reported on the synthesis, DNA complexation, and overall in vitro transfection efficiency of these conjugates. In this work, we characterize the intracellular trafficking of this system in order to identify the bottlenecks to more efficient delivery with these synthetic vectors, and further elucidate the mechanisms which enable any enhanced efficiency of this system over other common polyamines. We also introduce a new method for high-throughput screening of the endosomal escape properties of various polyplex formulations.
Materials and Methods
Materials
Generation 5.0 cystamine core PAMAM dendrimers were obtained from Dendritic Nanotechnologies (Mount Pleasant, MI) and used without further purification. Heterobifunctional poly (ethylene glycol) (Maleimide-PEG5k- N-hydroxysuccinimide) was obtained from Laysan Bio (Arab, AL). WIFPWIQL peptide was synthesized and purified by the MIT Biopolymers lab in the Swanson Core facilities at the Koch Institute. Immobilized Tris(2-carboxyethyl)phosphine (TCEP) reducing gel was obtained from Pierce (Thermo Fisher Scientific, Rockford, IL). Branched 25,000 g/mol (Mw) polyethyleneimine (PEI) and other chemical reagents were purchased from Sigma Aldrich (St. Louis, MO). DU145 cells and cell culture media were obtained from ATCC (Manassas, VA). pCMV-EGFP-N1 plasmid DNA was obtained from Aldevron Inc.(Fargo, ND). All other cell culture reagents were obtained from Invitrogen (Carlsbad, CA).
Block Copolymer Synthesis
PAMAM-G5-PEG block copolymers were prepared in a three-step synthesis. In the first step, generation 5.0 cystamine core PAMAM dendrimer (60.0 mg, 2.05 μmol) was reduced using immobilized TCEP gel (1.5 mL) for 90 min. Confirmation of the reduction was done using (5, 5′-dithiobis-(2-nitrobenzoic acid) (DTNB, Ellman’s reagent) to quantify free thiols and fluorescamine to quantify primary amines. Reduction was greater than 80%. In parallel, targeting peptide WIFPWIQL (7.1 mg, 6.5 μmol) was dissolved in DMSO at 5 mg/mL and added to a solution of Mal-PEG5k-NHS (30 mg, 5.4 μmol) in DMSO at 5 mg/mL. 20 μL triethylamine (TEA) was added to facilitate the reaction. After 30 min, this reaction was twice precipitated in cold ether, dissolved in PBS and reacted with the reduced PAMAM dendrimer for 24 hours. Untargeted polymers were synthesized by substituting monofunctional MAL-mPEG5k for the WIFPWIQL-PEG-MAL. The synthesized polymers were dialyzed against 8000 MWCO SpectraPOR (Spectrum Labs, Rancho Dominguez, CA) and lyophilized. Structure was confirmed by 1H-NMR (400MHz, D2O) δ (ppm) = 3.69 (s, CH2CH2O), 3.48 (t, CONHCH2CH2NH2), 3.30 (t, CONHCH2CH2NR), 3.11 (t, CONHCH2CH2NH2), 2.85 (m, NCH2CH2CONH), 2.64 (t, CONHCH2CH2NR), 2.45 (m, NCH2CH2CONH)
Cell Culture
DU145 cells were cultured in Minimum Essential Media (MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin in a humidified 37°C atmosphere at 5% CO2.
Transfection
Cells were trypsinized prior to transfection and seeded in 96-well plates at 5,000 cells/well overnight in a humidified 37°C atmosphere at 5% CO2. PAMAM-PEG block copolymers were diluted in 25 mM sodium acetate buffer (NaAc) (pH 5.5) at 1.5 mg/mL initially and then further diluted into 25 μL of the same buffer in a separate 96-well plate at varying concentrations. 25 μL of pDNA at 0.06 mg/mL in NaAc was then added to each well and mixed by gentle pipetting. After 10 min for complex formation, 30 μL of the complexes were added to 200 μL of Opti-MEM supplemented with 10% FBS. After further mixing, 150 μL of the complexes in Opti-MEM were added to the cells (after growth media had been removed). After 4 hours, the complexes in Opti-MEM were removed and growth media was added. Cells were assayed at 48 hr for GFP production using flow cytometry. 4000 – 10000 live cells per sample were analyzed. GFP production was normalized to that from transfection with an optimized formulation of 25kDa branched PEI (30). p-values were calculated using a two-way analysis of variation and Bonferroni post-tests were performed on all conditions shown.
Flow Cytometry
Flow cytometry was performed in U-bottom 96-well plates using an LSR II HTS Flow cytometer (Becton-Dickinson, Mountain View, CA). To prepare samples, media was removed from cells and replaced with 25 μL trypsin for 5 minutes. 50 μL of PBS supplemented with 2% FBS was then added to each well, mixed, and the entire 75 μL cell suspension transferred into a U-bottom 96-well plate. Representative 2D histograms are included in the Supporting Information.
Polyplex Uptake
Block copolymers and PEI were labeled with fluorescein isothiocyanate (FITC) at a molar ratio of 4:1 (dye:polymer). Using the labeled polymers, polyplexes were formed and cells treated as described in the above section. At the time indicated, cells were removed from the incubator and analyzed using flow cytometry. p-values were calculated using a two-way analysis of variation and Bonferroni post-tests were performed on all conditions shown.
High-Throughput Endosomal Escape
Complexes were assembled and transfection was conducted as described above, except that 25 μM calcein was added to the Opti-MEM and cells were seeded in black-sided, clear-bottom 96-well plates. 4 hours after transfection, 5 μL of a solution of Hoechst 33342 diluted to 1:30 in PBS was added. After 20 minutes of staining, complexes and free dye were removed, and the cells were washed 3 times with PBS. 150 μL of phenol-free Opti-MEM with 10% serum was added to each well before the plate was covered with an opaque sticker, foiled, and analyzed. Imaging was done using a Cellomics ArrayScan VTI HCS Reader (Thermo Fisher, Waltham, MA) and analysis was done using the included software (see Supporting information for details). 300 – 3000 live cells per sample were analyzed. p-values were calculated using an unpaired t-test.
Confocal Microscopy
8-well Lab-Tek chamber slides (Thermo Fisher, Waltham, MA) were treated for 20 minutes with human fibronectin in PBS at 0.01 mg/mL. The fibronectin was removed and DU145 cells were trypsinized and seeded in each well at a concentration of 1000 cells/well 24 h before transfection. Plasmid DNA was labeled with Rhodamine-CX using the Label-IT kit (Mirus Bio, Madison, WI). Polymers were FITC-labeled as described earlier. Polyplexes between labeled DNA and labeled polymers were formed in NaAc buffer as described earlier. 40 μL of complexes were added to 160 uL phenol-free Opti-MEM (supplemented with 10% serum) and added to each well. Complexes were removed after 4 h and replaced with growth media. At 24 h, cells were fixed with 3.7% formaldehyde in PBS, stained with Hoechst 33342, and were washed 3 times with PBS. Imaging was done on a PerkinElmer Ultraview spinning disc confocal (PerkinElmer, Waltham, MA).
Results and Discussion
Synthesis
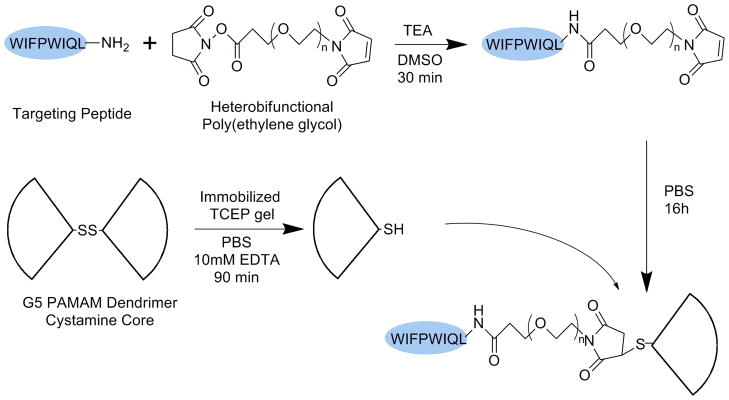
Synthesis of the PAMAM-G5-PEG linear-dendritic block copolymers was similar to that reported previously by our group (26) and is shown in Scheme 1. Briefly, generation 5.0, cystamine core dendrimers were first reduced to expose dendrons with a single thiol moiety (yielding two dendron-thiols per dendrimer). In parallel, the N-hydroxysuccinimide (NHS) group of a heterobifunctional PEG was first reacted with the terminal lysine on the WIFPWIQL peptide, leaving the maleimide group of the PEG block free for subsequent conjugation to the exposed thiol on the reduced PAMAM dendrimer. This synthetic approach is managed easily in aqueous solutions and can be generally applied to most targeting ligands.
Scheme 1.
Synthesis of PAMAM-G5-PEG-WIFP conjugates. The targeting peptide is first reacted with a bifunctional linker before being precipitated and added to reduced G5 PAMAM dendrimer.
Overall Transfection
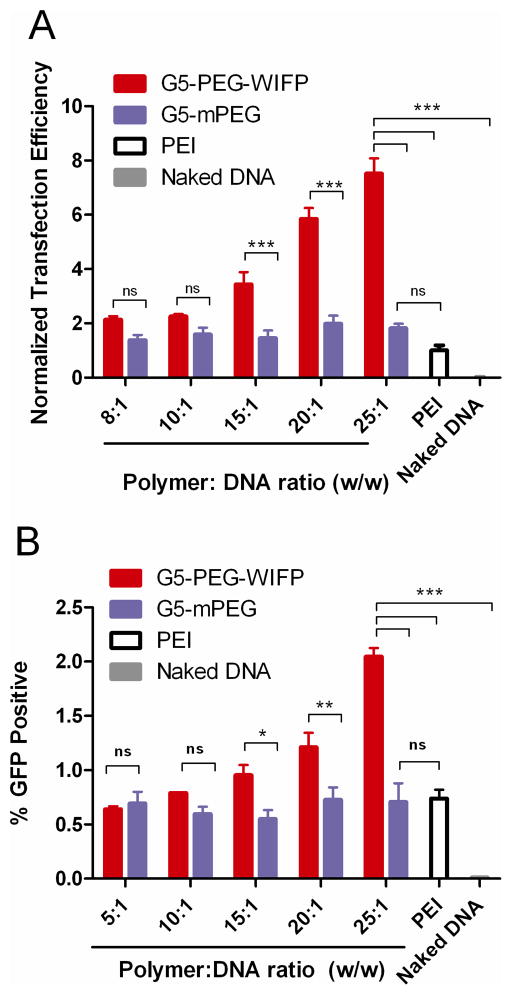
While the overall transfection efficiency of a gene carrier is a critical parameter to characterize, it ultimately necessary to determine the bottlenecks – quantitatively if possible – in order to better design future iterations of the system. As shown in Figure 1A, the targeted block copolymers transfected DU145 cells nearly 8-fold better than 25kDa bPEI and nearly 4-fold better than an untargeted control block copolymer, which is consistent with our previously published studies using this system (26). Figure 1B show that the presence of serum in the system limited the absolute percentage of cells transfected to a less than 3% and the fold increase of targeted block copolymer was 3-fold over both the untargeted control and PEI. While PEI is an efficient gene carrier in serum-free media, in more biologically relevant conditions (i.e. 10% or more serum), the advantage of the PEGylation in the block copolymer system becomes apparent (31).
Figure 1.
Normalized transfection efficiency (A) and percentage of cells transfected (B) of targeted and untargeted block copolymer formulations at various polymer:DNA ratios. Transfections of EGFP were performed in 10% serum and analyzed 48 hrs later using flow cytometry. An optimized formulation of PEI at a polymer:DNA ratio of 2:1 was used and data is reported as mean fluorescence intensity normalized to that of the PEI transfected cells. Error bars represent SEM of 5 replicate experiments (*** = p < 0.001, ** = p < 0.01, * = p < 0.05, ns = not significant). These results are in agreement with previously published reports of transfection efficiency of this block copolymer system (26).
Uptake
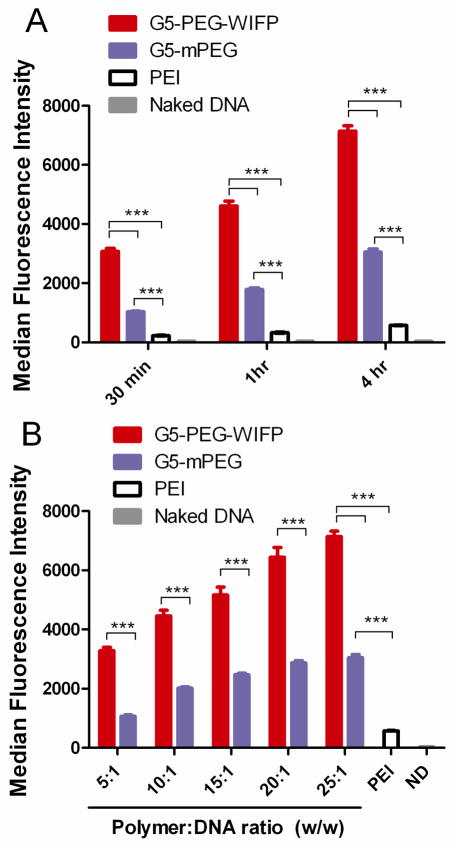
Once in the immediate vicinity of the target cell, the polyplex must first undergo binding and internalization. While it is possible to characterize many aspects of the binding and internalization process (32), at this time we chose to simply focus on the degree of polyplex association with the target cells. In this study, each polymer system was labeled with a fluorophore (FITC) and the labeled polyplexes were added to DU145 cells. Figure 2 shows the median cell associated fluorescence achieved by the various delivery vehicles after 1–4 hrs of incubation, as measured by flow cytometry. Intracellular uptake of the PAMAM-G5-PEG WIFP block copolymer complexes exhibit a 12-fold increased cellular uptake over PEI; the untargeted PAMAM-G5-PEG also yielded a 5-fold gain versus the PEI control. This is consistent with other reports that the PEG exterior shell generated in the block copolymer polyplexes gives a significant advantage in shielding them from adsorption of proteins (33). The presence of serum proteins (particularly albumin) can abrogate the uptake of PEI by binding PEI complexes prior to cell uptake (31). Of note is that the general relationship between the uptake efficiencies and overall transfection efficiencies of these polymers is similar, implying that at least one principal difference between their effectiveness is their differential uptake ability. However, the fold increases in the percentage of transfected cells of the block copolymers over PEI are much less than the fold increases in uptake, suggesting that PEI may be more effective at mediating downstream events. Another difference could lie in the endosomal trafficking of polyplexes internalized via GRP78 binding (34) versus those internalized via non-receptor mediated endocytosis, as increased uptake resulted in increased transfection efficiency for the targeted polymer, but not for the untargeted control. This effect could also be due to potential downstream effects of the peptide in mediating endosomal escape or nuclear translocation. For each of the polymers, even though significant uptake is achieved, further evaluation is necessary to understand the subsequent barriers to transfection that result in relatively low transgene expression in comparison to viral vectors. A block copolymer with a scrambled peptide may have been a more ideal targeting control; however the polyplex size, charge and loading are unchanged by the presence of the peptide, indicating that the PEG control should be a good control of a non-targeted system (see Supporting information). Additionally, our earlier communication (26), shows that the addition of competing ligand suppresses overall transfection efficiency, indicating that the uptake of these particles is receptor-mediated.
Figure 2.
Uptake of polyplexes formed by targeted and untargeted block copolymers against PEI. FITC-labeled polymers were complexed with EGFP plasmids and added to DU145 cells in 10% serum. Median cell associated fluorescence was measured at 30 min, 1h, and 4h as indicated in panel A (w/w = 25:1 for block copolymers). Panel B shows the effect of varying the polymer:DNA ratio of the block copolymers. Fluorescence was measured at 4 h in panel B. PEI was used at an optimized w/w ratio of 2:1. Error bars represent SEM of 5 replicate experiments (*** = p < 0.001, ** = p < 0.01, * = p < 0.05, ns = not significant).
Endosomal Escape
Endosomal escape is a critical barrier to overcome in the delivery of nucleic acids. Measurement of endosomal escape is done frequently using fluorescence microscopy (35). Commonly, a fluorescent marker for the endosomes (transferrin, fluorophore-modified dextrans, endosome/lysosome tracking dye) is employed, and escape is considered achieved if there is no co-localization between the polyplex and the endosomal marker (36). However, this is difficult to measure and quantify in high throughput, thus screening a large library of materials or formulations for differences in endosomal escape performance is complex. Akinc et al. (37) describe a novel method for measuring the pH of the polyplex environment via high-throughput flow cytometry, but here we describe a direct method for quantifying the disruption of the endosome using an image-based high-throughput screening approach.
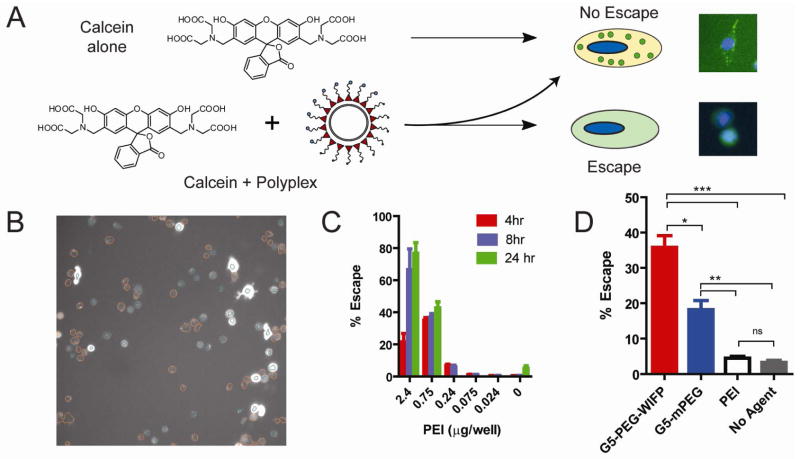
The high-throughput approach uses the fact that calcein is a membrane-impermeable fluorophore that can be taken up by cells and trafficked through endosomes to lysosomes, but cannot achieve endosomal/lysosomal escape (38, 39). In the presence of an escape agent, calcein will be released and is seen throughout the cell. (Fig. 3A). Using an automated fluorescent microscope, the two phenotypes can be differentiated in real-time by algorithms using spatial fluorescence information. Fig. 3B shows a representative image (20x magnification) in which the algorithm has identified cells which have undergone endosomal escape vs. those that have not. Figure 3C is an example of an assay validation experiment in which increasing amounts of PEI were added and the escape measured at different time points. As expected, escape percentage increased with increasing concentrations of PEI and increasing incubation time. When applied to the block copolymer systems studied, a similar pattern to that of the uptake and overall transfection efficiency was observed. The observed escape percentage relationship between targeted and untargeted PAMAM block copolymers was similar to that of the uptake, suggesting the endosomal escape properties were relatively unchanged between the two polymers. This is consistent with the fact that these two polymer differ only in the presence of a targeting ligand, and thus the pH-responsive PAMAM structure remains the same. The low escape percentage of the PEI is likely due to its poorer uptake relative to the block copolymers as its escape percentage is greatly enhanced in the absence of serum (see Supporting Information).
Figure 3.
Panel A is a schematic of the high-throughput calcein assay used to quantify endosomal escape. When calcein alone is taken up by cells, it remains in the endosomal compartments and thus a punctuate pattern of fluorescence in the cytoplasm is visible. With the addition of an endosomal escape agent, e.g. a polymer or polyplex, the calcein is released into the cytosol yielding a uniform pattern of fluorescence. The images in panel A are representative of these morphologies and taken at 20x. The images have been intensity normalized for this figure in order to more easily visualize the punctuate vs diffuse pattern of fluorescence. Nuclei are shown in blue, calcein as green. Panel B is a grayscale image of the calcein channel of DU145 cells treated with G5-PEG-WIFP polyplexes. The Cellomics algorithm has differentiated cells with endosomal escape (blue circle overlay) vs. those without (orange circle overlay). In panel C, the concentration of PEI is increased as is the time in which the cells were exposed to the calcein/escape agent. Panel D shows the percentage of escaped cells from treatments with various polyplex formulations in OPTI-MEM with 10% serum. PAMAM based polyplexes were used at a 25:1 w/w ratio. PEI was used at 2:1 w/w ratio. Error bars are SEM of 3 replicates (*** = p < 0.001, ** = p < 0.01, * = p < 0.05, ns = not significant).
Nuclear Uptake and DNA Unpackaging
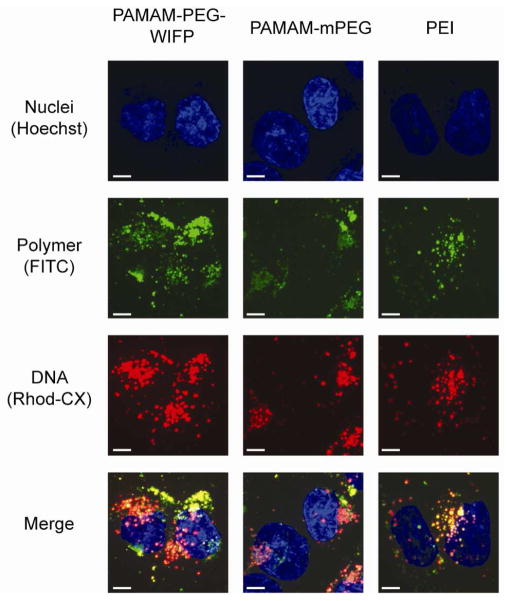
While endosomal escape could be visualized and quantitated using a high-throughput approach, dissociation of the polyplex and nuclear uptake were followed using traditional confocal microscopy techniques. In this case, both the plasmid DNA and polymers were labeled in order to track polyplex dissociation. Figure 4 shows representative images of a 24-hour incubation of dual-labeled polyplexes. There is substantially more plasmid DNA delivered by the targeted block copolymer in comparison with the untargeted polymer or PEI. Again, this is due primarily to the differential uptake between the three systems. Additionally, it is apparent that the PAMAM-PEG micelles are not completely dissociated, as there is significant co-localization between the polymer and DNA channels. In the case of the targeted PAMAM-PEG, it is possible to visualize a yellow core at the center of areas of red fluorescence, indicating that there is still polymer associated with most polyplexes. Finally, it is evident that the polyplexes are localized to an area near the nucleus, but not inside, such as a microtubule organization center (MTOC) (40). Uptake into the nucleus should be readily seen 24 hours after initial application, but the fact that it is not observed with these materials indicates nuclear localization to be a significant barrier to delivery. The observed gene expression can likely be attributed to either the small percentage of decomplexed DNA that is non-specifically chaperoned into the nucleus or to cells in which the nuclear membrane was not intact (e.g. in the process of dividing) (41). In this case, some polyplexes accessing the nucleus may still be fully or partially condensed, but even partially complexed polyplexes could participate in gene transcription (42). The lack of efficient nuclear uptake and polyplex decondensation represents a significant barrier to further transfection efficiency and a represents an area in which these polymers can be improved, possibly by the incorporation of a nuclear localization sequence.
Figure 4.
Confocal fluorescence micrographs of cells transfected with targeted/untargeted block copolymers as well as PEI. Transfection took place in 10% serum and micrographs were taken on fixed cells 24h after polyplexes were added. Scale bars are 5 microns. In the merged channel, yellow indicates colocalization of polymer (green) and DNA (red).
For the targeted block copolymers, nearly 100% of cells took up complexes (see Supporting Information), 36% achieved endosomal escape, but only 2% were ultimately transfected. Less than 6% of cells which experienced escape were transfected by either targeted or untargeted block copolymer, while over 15% of escaped cells were transfected by PEI. This underscores that while the escape of these block copolymers polymers is efficient, downstream events after endosomal escape represent the most important challenges going forward. Addressing these challenges will be important because even as currently designed, these promising targeted block copolymers are capable of superior transfection relative to PEI in the presence of serum.
Conclusions
PAMAM-PEG block copolymers have shown promise as gene delivery agents. Here we have shown this system to be nearly an order of magnitude more potent than PEI in vitro and have examined the bottlenecks the system faces. While targeted uptake remains efficient, even in the presence of serum, endosomal escape and unpackaging of DNA polyplexes afterward could be further improved. To that end, we have demonstrated a tool to screen libraries of compounds to rapidly determine the best structures and formulations for endosomal escape. Finally, we have shown that nuclear translocation appears to be the primary obstacle to more efficient transfection in this system – further engineering of the system to promote active delivery of polyplexes into the nucleus could greatly enhance transfection.
Supplementary Material
Acknowledgments
Funding for this research was provided by National Institutes of Health (NIH) NIBIB grant R01EB008082. This work was also supported by National Defense Science and Engineering Graduate Fellowship and the National Science Foundation Graduate Research Fellowship Program, as well as The MIT-Harvard Center of Cancer Nanotechnology Excellence Center, NCI grant 1U54CA151884. The authors acknowledge the Swanson Biotechnology Center in the Koch Institute at MIT, the Keck Imaging facility at the Whitehead Institute, and the Genome Technology Core at the Whitehead Institute. The authors also thank James Evans, Roberto Zoncu, and Nicki Watson for helpful imaging advice and assistance.
References
- 1.Somia N, Verma IM. Gene therapy: trials and tribulations. Nat Rev Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 2.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene therapy. 2008;15:1131–8. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 3.Walther W, Stein U. Viral vectors for gene transfer - A review of their use in the treatment of human diseases. Drugs. 2000;60:249–271. doi: 10.2165/00003495-200060020-00002. [DOI] [PubMed] [Google Scholar]
- 4.Zaiss AK, Muruve DA. Immunity to adeno-associated virus vectors in animals and humans: a continued challenge. Gene Ther. 2008;15:808–816. doi: 10.1038/gt.2008.54. [DOI] [PubMed] [Google Scholar]
- 5.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Therapy. 2010;17:295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glover DJ, Lipps HJ, Jans DA. Towards Safe, Nonviral Therapeutic Gene Expression in Humans. Nature Reviews Genetics. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 7.Kan PL, Schatzlein AG, Uchegbu IF. Polymers Used for the Delivery of Genes in Gene Therapy. In: Uchegbu IF, Schatzlein AG, editors. Polymers in Drug Delivery. CRC Press; Boca Raton: 2006. [Google Scholar]
- 8.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofection - a Highly Efficient, Lipid-Mediated DNA-Transfection Procedure. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kogure K, Akita H, Yamada Y, Harashima H. Multifunctional envelope-type nano device (MEND) as a non-viral gene delivery system. Advanced Drug Delivery Reviews. 2008;60:559–571. doi: 10.1016/j.addr.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Legendre JY, Szoka FC., Jr Delivery of Plasmid DNA into Mammalian Cell Lines Using pH-Sensitive Liposomes: Comparison with Cationic Liposomes. Pharmaceutical Research. 1992;9:1235–1242. doi: 10.1023/a:1015836829670. [DOI] [PubMed] [Google Scholar]
- 11.Haensler J, Szoka FC. Polyamidoamine Cascade Polymers Mediate Efficient Transfection of Cells in Culture. Bioconjugate Chemistry. 1993;4:372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 12.Tang MX, Redemann CT, Szoka FC. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chemistry. 1996;7:703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 13.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Human Gene Therapy. 1996;7:1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 14.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol Ther. 2005;11:426–34. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed Engl. 2003;42:3153–8. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 16.Lynn DM, Anderson DG, Putnam D, Langer R. Accelerated discovery of synthetic transfection vectors: parallel synthesis and screening of a degradable polymer library. J Am Chem Soc. 2001;123:8155–6. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 17.Fukushima S, Miyata K, Nishiyama N, Kanayama N, Yamasaki Y, Kataoka K. PEGylated Polyplex Micelles from Triblock Catiomers with Spatially Ordered Layering of Condensed pDNA and Buffering Units for Enhanced Intracellular Gene Delivery. Journal of the American Chemical Society. 2005;127:2810–2811. doi: 10.1021/ja0440506. [DOI] [PubMed] [Google Scholar]
- 18.Chiou HC, Tangco MV, Levine SM, Robertson D, Kormis K, Wu CH, Wu GY. Enhanced Resistance to Nuclease Degradation of Nucleic-Acids Complexed to Asialoglycoprotein-Polylysine Carriers. Nucleic Acids Research. 1994;22:5439–5446. doi: 10.1093/nar/22.24.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dash PR, Read ML, Barrett LB, Wolfert M, Seymour LW. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Therapy. 1999;6:643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]
- 20.Patel HM. Serum opsonins and liposomes: their interaction and opsonophagocytosis. Crit Rev Ther Drug Carrier Syst. 1992;9:39–90. [PubMed] [Google Scholar]
- 21.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam AP, Dean DA. Progress and prospects: nuclear import of nonviral vectors. Gene therapy. 2010:1–9. doi: 10.1038/gt.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean DA, Strong DD, Zimmer WE. Nuclear entry of nonviral vectors. Gene Therapy. 2005;12:881–90. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hama S, Akita H, Iida S, Mizuguchi H, Harashima H. Quantitative and mechanism-based investigation of post-nuclear delivery events between adenovirus and lipoplex. Nucleic acids research. 2007;35:1533–43. doi: 10.1093/nar/gkl1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood KC, Little SR, Langer R, Hammond PT. A family of hierarchically self-assembling linear-dendritic hybrid polymers for highly efficient targeted gene delivery. Angewandte Chemie (International ed in English) 2005;44:6704–8. doi: 10.1002/anie.200502152. [DOI] [PubMed] [Google Scholar]
- 26.Wood KC, Azarin SM, Arap W, Pasqualini R, Langer R, Hammond PT. Tumor-targeted gene delivery using molecularly engineered hybrid polymers functionalized with a tumor-homing peptide. Bioconjugate chemistry. 2008;19:403–5. doi: 10.1021/bc700408r. [DOI] [PubMed] [Google Scholar]
- 27.Arap MA, Lahdenranta J, Mintz PJ, Hajitou A, Arap W, Pasqualini R. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell. 2004;6:275–284. doi: 10.1016/j.ccr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Lee AS. GRP78 Induction in Cancer: Therapeutic and Prognostic Implications. Cancer Research. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Lee AS. Stress Induction of GRP78/BiP and Its Role in Cancer. Current. 2006:45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- 30.Gabrielson NP, Pack DW. Acetylation of Polyethylenimine Enhances Gene Delivery via Weakened Polymer/DNA Interactions. Biomacromolecules. 2006;7:2427–2435. doi: 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]
- 31.Forrest ML, Meister G, Koerber J, Pack D. Partial Acetylation of Polyethylenimine Enhances In Vitro Gene Delivery. Pharmaceutical Research. 2004;21:365–371. doi: 10.1023/b:pham.0000016251.42392.1e. [DOI] [PubMed] [Google Scholar]
- 32.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. Journal of Controlled Release. 145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giri J, Diallo MS, Simpson AJ, Liu Y, Goddard WA, Kumar R, Woods GC. Interactions of Poly(amidoamine) Dendrimers with Human Serum Albumin: Binding Constants and Mechanisms. ACS Nano. :Article ASAP. doi: 10.1021/nn1021007. [DOI] [PubMed] [Google Scholar]
- 34.Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434:181–8. doi: 10.1042/BJ20101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne CK. Imaging gene delivery with fluorescence microscopy. Nanomedicine. 2007;2:847–60. doi: 10.2217/17435889.2.6.847. [DOI] [PubMed] [Google Scholar]
- 36.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative Three-Dimensional Analysis of the Intracellular Trafficking of Plasmid DNA Transfected by a Nonviral Gene Delivery System Using Confocal Laser Scanning Microscopy. Mol Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. The journal of gene medicine. 2005;7:657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 38.Hu Y, Litwin T, Nagaraja AR, Kwong B, Katz J, Watson N, Irvine DJ. Cytosolic Delivery of Membrane-Impermeable Molecules in Dendritic Cells Using pH-Responsive Core-Shell Nanoparticles. Nano Letters. 2007;7:3056–3064. doi: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]
- 39.Jones RA, Cheung CY, Black FE, Zia JK, Stayton PS, Hoffman AS, Wilson MR. Poly(2-alkylacrylic acid) polymers deliver molecules to the cytosol by pH-sensitive disruption of endosomal vesicles. Biochemical Journal. 2003;372:65–75. doi: 10.1042/BJ20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barua S, Rege K. The influence of mediators of intracellular trafficking on transgene expression efficacy of polymer-plasmid DNA complexes. Biomaterials. 31:5894–5902. doi: 10.1016/j.biomaterials.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Pouton CW, Wagstaff KM, Roth DM, Moseley GW, Jans DA. Targeted delivery to the nucleus. Advanced Drug Delivery Reviews. 2007;59:698–717. doi: 10.1016/j.addr.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Bielinska AU, Kukowska-Latallo JF, Baker JR. The interaction of plasmid DNA with polyamidoamine dendrimers: mechanism of complex formation and analysis of alterations induced in nuclease sensitivity and transcriptional activity of the complexed DNA. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1997;1353:180–190. doi: 10.1016/s0167-4781(97)00069-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.