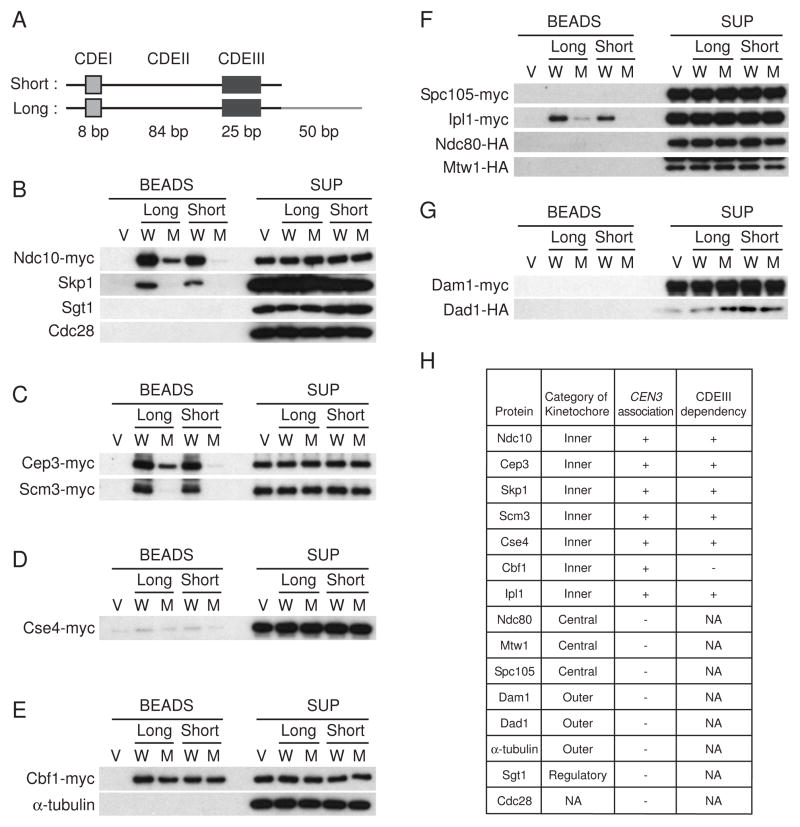
Figure 1. Initial kinetochore assembly in vitro.
(A) The 117-bp CEN3 DNA consists of three conserved DNA elements, designated CDEI, CDEII, and CDEIII. CDEI consists of a conserved 8-bp sequence incorporating a 6-bp palindrome, which is important for Cbf1 association. Approximately 93% of the CDEII sequence is made up of A:T base pairs. CDEIII has a size of 25 bp and includes the completely conserved sequence CCG. As a negative control in the in vitro kinetochore assembly system, the CCG sequence of CDEIII was replaced to CCC. The short version of CEN3 DNA has a length of 134 bp, containing only CDEI, CDEII, and CDEIII. The long version of CEN3 DNA has a length of 184 bp, containing the same sequences plus an Ndc10-binding site (see Espelin et al., 1997 [18]). (B–G) Protein extracts from cyclic cells were incubated with magnetic beads coupled to plasmid DNA. Vector (V) or plasmid containing 8 tandem copies of wild-type (W) or mutant (M) CEN3 was used. After 4 min at room temperature with highly concentrated protein extract, the beads were collected and washed (Figure S1B). Proteins bound to the beads were analyzed by 4–15% SDS-PAGE and immunoblotting. The protein extracts derived from the tagged strains were as follows: (B) Ndc10-myc Ndc80-HA Mtw1-HA (Y2046); (C) Cep3-myc Scm3-myc Mif2-myc (Y2049); (D) Cse4-myc (Y2044); (E) Cbf1-myc Isw1-HA (Y2083); (F) Spc105-myc Ipl1-myc Ndc80-HA Mtw1-HA (Y2047); and (G) Dam1-myc Stu2-myc Dad1-HA (Y2048). (H) Summary of kinetochore assembly in 4 min. NA: Not applicable.