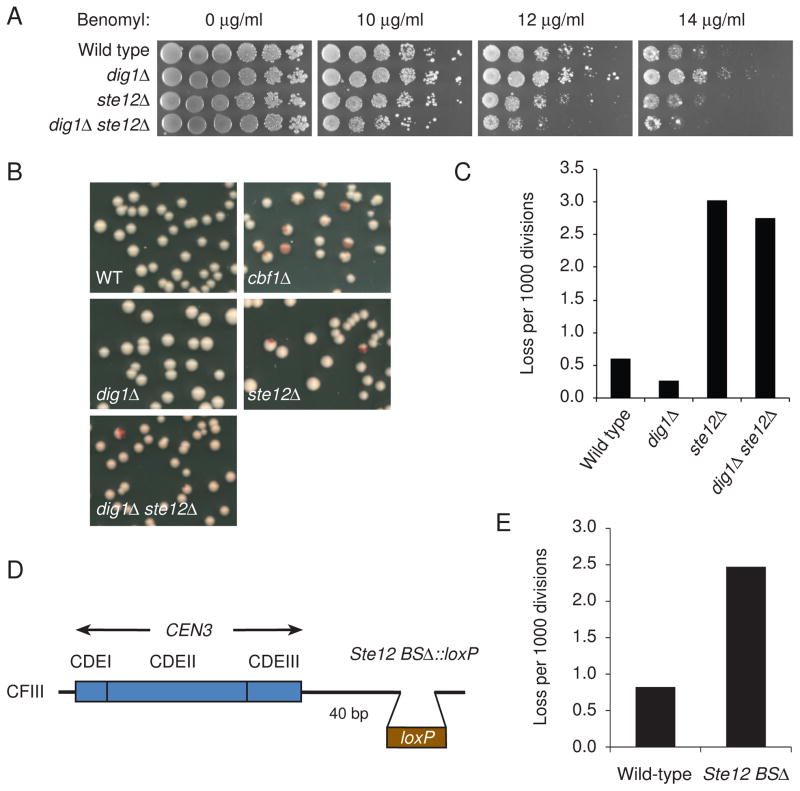
Figure 4. Centromeric transcripts contribute to centromere function.
(A) Benomyl resistance of dig1Δ cells and benomyl sensitivity of ste12Δ and dig1Δ ste12Δ cells. Yeast cells were spotted in 5-fold dilutions from 5 × 104 cells per spot on YPD plates containing benomyl. The plates were incubated at 23°C for 5 days and photographed. The yeast strains used were as follows: wild-type (YPH499), dig1Δ (Y1978), ste12Δ (Y1985), and dig1Δ ste12Δ (Y2056). (B) cbf1Δ, ste12Δ, and cbf1Δ ste12Δ mutants sectoring phenotypes. Each strain includes a single SUP11-marked chromosome III fragment containing CEN3. (C) Chromosome loss rate in null mutants was determined by half-sector analysis. Wild type: 18 half-sectored colonies/30,214 total colonies; dig1Δ: 6/22,972; ste12Δ: 44/14,567; dig1Δ ste12Δ: 40/14,575. The yeast strains used in B and C were as follows: wild-type (Y14), cbf1Δ (Y2011), dig1Δ (Y2016), ste12Δ (Y2060 and Y2061), and dig1Δ ste12Δ (Y2062 and Y2063). (D) Schematic diagram showing deletion of the Ste12 binding site (Ste12 BSΔ) on the chromosome fragment. (E) Increased chromosome missegregation in Ste12 BSΔ. Chromosome loss rate was determined by half-sector analysis. Wild-type (Y14): 7 half-sectored colonies/8,550 total colonies; Ste12 BSΔ (Y2082): 20/8,085.