Abstract
The prevalence of obesity and diabetes has increased exponentially in recent years around the globe, especially in India. Sweet proteins have the potential to substitute the sugars, by acting as natural, good and low calorie sweeteners. They also do not trigger a demand for insulin in diabetic patients unlike sucrose. In humans, the sweet taste perception is mainly due to taste-specific G protein-coupled heterodimeric receptors T1R2-T1R3. These receptors recognize diverse natural and synthetic sweeteners such as monelin, brazzein, thaumatin, curculin, mabinlin, miraculin and pentadin. Structural modeling of new sweetener proteins will be a great leap in further advancement of knowledge and their utility as sweeteners. We have explored the fingerprints of sweetness by studying the aminoacid composition and structure properties of the above proteins. The structural analysis of monellin revealed that the individual A or B chains of monellin are not contributing to its sweetness. However, the native conformation and ionic interaction between AspB7 of monellin with active site of T1R2-T1R3 receptor, along with hydrogen bonding stability of IleB6 and IleB8 are responsible for the sweet taste. Based on structural similarity search, we found a new hypothetical protein from Shewanella loihica, which has the presence of Asp32 with adjacent isoleucine residues. Further, we examined the lead protein by two-step docking for the study of interaction of functionally conserved residues with receptors. The identified protein showed similar ionic and hydrophobic interactions with monelin. This gives a promising opportunity to explore this protein for potential health application in the low calorie sweetener industry viz., soft drinks, snacks, food, chocolate industries etc.
Keywords: sweet protein, Shewanella loihica, taste receptor
Background
The prevalence of obesity and diabetes has increased exponentially in recent years around the globe, with special mention about India. Sweet proteins have the potential to replace the sugars by acting as natural, good, low-calorie sweeteners. The design of safer sweeteners is very important for people who are affected by diabetes, hyperlipidemia and other diseases that are linked to the consumption of sugars. Sweet proteins which are found in several tropical plants, are many times sweeter than sucrose on a molar basis [1]. Although sugar substitute are now reasonably safer, there is a continuous search for new, and safer ones. Their design requires a good understanding of the interaction of sweetener molecules with their receptors [2]. The physiological basis of sweet taste perception has captured the imagination of many philosophers, cooks and scientists [3]. A good understanding of their structure-function relationship can compliment SAR studies on small molecular weight sweeteners and thus help in the design of safe sweeteners [1]. Research groups have recently identified a likely sweet taste receptor, T1R3 [4]. Despite numerous studies on taste receptors there remains a great deal of understanding in what characteristics of the molecules govern the perception of taste [5]. The computational studies on molecular recognition between proteins and low molecular weight molecules have experienced a significant development in the recent decade. Various enzyme and receptor systems of chemical, biological and medicinal interest have been studied by simulating ligand-protein interactions at the molecular level [6]. These computational approaches give a promising vision to predict, find and explore the availability of new sweetener products especially low-molecular proteins, non-conventional natural plants and microbial sources. Previous studies show that there is virtually no homology among the known sweet proteins, but a little structural similarity exists. In monelin (209U) replacement of AspB7 by L-2 aminobuteric acid, Gly and D-Asp resulted in complete loss of sweetness. Replacement of IleB6 and IleB8 by different amino acids resulted in significant decrease of sweetness, or complete loss of sweetness. These results suggested that the free β-carboxyl group of the AspB7 would possibly bind to the receptor site through ionic bonding and trigger the sensation of intense sweet taste, and IleB6 and/or IleB8 would be involved in the hydrophobic interaction with the receptor site [7]. Similarly our detailed study also stands with slight and very vital different that the Ile residues making hydrophobic interactions with residues in the active sites of receptor protein. The binding regions are conserved in most of them and they posses similar secondary structure formation and relative reasonable tertiary structure in monelin, curculin and mablin. Residues like IleB6, AspB7 and IleB8 in monelin identical residues are there in curculin(2PDF) they are Ile36, Asp37 and Val38 here valaine is notable and Isoleusine are functionally identical secondary structure formation in protein folding. Similar structure search and structure comparison resulted in a hypothetical protein (2GPI), from a marine bacterium, Shewanella loihica. It also showed the availability of two adjacent residues, Ile5 and Ile6, and the formation of similar secondary structure suggests strongly the possibility of a sweet protein.
Methodology
Interaction of sweet fingers with T1R3 active site is carried out by docking studies. Since T1R3 is a membrane protein, it has not been crystallized and its structure is not yet determined. To perform virtual interaction studies T1R3 structure was developed by homology modeling. Previous studies on homology modeling of T1R3 protein used 1EWK [8] and 1EVW [9] as template to build the extra-cellular domains and/or complete backbone. The IEWK shows 27% of sequence identity and 35% of similarity, and the 1EVW shows 40% sequence identity with T1R3 amino acid sequence. In the modeling approach, we used 2E4U as template protein which is a crystal structure of extra cellular region of the metabotropic glutamate receptor and showed 75% sequence similarity. SWISS-PDB viewer and workspace are used for model building and energy refinement. Energy minimization was performed up to 20 times to get a biologically stabilized conformation. The T1R3 receptor model was validated using Ramachandran Plot available in CCP4 computational crystallographic suite [10]. It showed 85% of residues are in the favored region, 10% of the residues in the allowed region and remaining 5% of residues also in outlier region (Figure 1).
Figure 1.
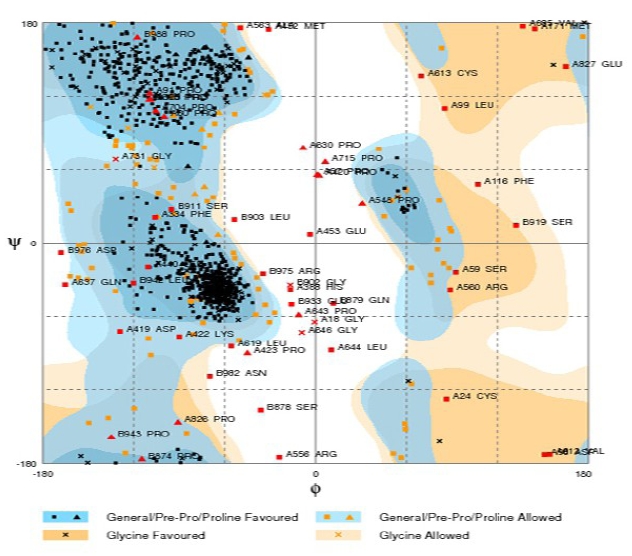
Ramachandran Plot showing residues in the optimal energy level.
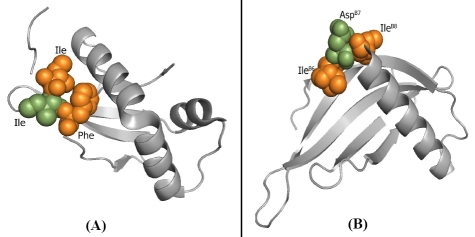
The relationship between amino acid sequence and structure gives deeper insights into the basic principles of protein structure and function. Study of monelin, an intensely sweet protein reported that some structural domains are working as sweet fingers [7]. We considered that the free β-carboxyl group of certain Asp residue in monelin would possibly bind to the receptor site through ionic bonding and trigger the sensation of an intense sweet taste. Monelin was taken as query to search protein having similar structural properties with expected sweet fingers, likely present in this known protein. Search performed against monellin (209U) on ProteinDBS, showed a hypothetical protein from Shewanella loihica having reasonable structure similarity, among the top scored hits for 2GPI, with expected adjacent Ile residues assumed as sweet fingers. 2GPI is a conserved hypothetical single chain small protein with the length of 91 amino acids. Pairwise structural alignment of 2GPI with monelin gives high similarity (Figure 2A, 2B). Potently sweet compounds possess a hydrophobic site, which participates in the binding to the receptor through a hydrophobic interaction. In aspartyl peptide sweeteners, both the amino acids adjacent to Asp residue are restricted in shape, size, stereo chemistry, and so on [7].
Figure 2.
(A) Predicted novel sweet protein, Ile residue in green colour makes ionic interaction; Ile and Phe residues in orange colour make hydrophobic interactions with taste receptor. (B) Known intense sweet protein monelin (209U) with similar structural finger print.
The exact mechanism of sweet proteins with the T1R2-T1R3 sweet taste receptor had not yet been elucidated [2]. Interaction study of our modeled structure of T1R2-T1R3 protein and assumed novel sweet protein candidate 2GPI reveals the possibility of having active sites with sweet taste activity in the hypothetical protein. A new drug/molecule should fit the active site of a specific receptor. Although electrostatic, hydrophobic and van der Waals interactions affect greatly the binding affinity of the molecules, shape complementarity is a necessary condition. [11]. Protein-protein docking was carried out to examine the interaction between predicted active site of 2GPI and taste receptor protein by using PATCHDOCK [12]. Standard parameters with clustering RMSD value of 4.0 and default complex type were followed in the docking process. The final docking results were analyzed. Among the top docking solutions the complex with most optimal interaction distance was used to visualize the hydrophobic interaction (Figure 3).
Figure 3.
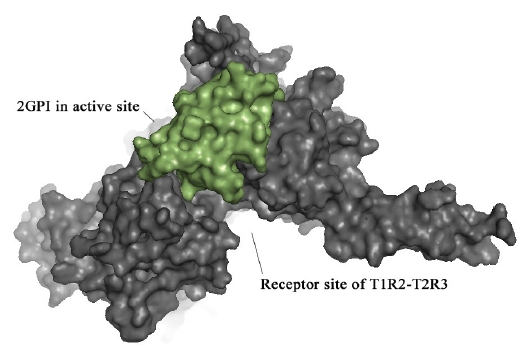
Dockpose of 2GPI in the active site of the extracellular region of T1R2-T1R3 receptor model.
Results
Computationally predicted hypothetical protein candidate is validated by protein-protein docking. The adjacent hydrophobic residues cause hydrophobic interaction with receptor site. One Asp residue between the hydrophobic amino acids (Figure 4) triggers ionic interaction with the active site of taste receptor protein as reported in previous studies. The 2GPI is a low molecular weight (10.5 K Da) protein. In addition the source of this hypothetical protein is a cultivatable marine bacterium, Shewanella loihica. Our study gives a promising opportunity to design and develop novel intense sweetener proteins through a structure-based similarity search and computational modeling techniques. It may ensure potential health beneficial sweetener products to enhance industrial production and marketing of low-calorie food items.
Figure 4.
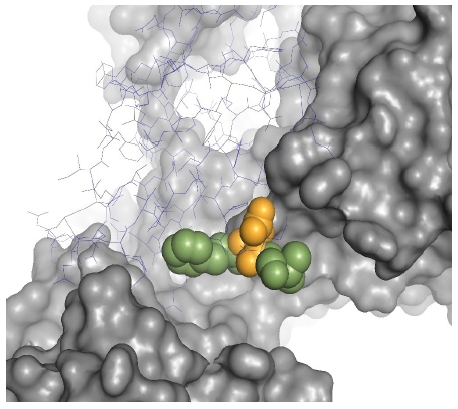
Residues making ionic and hydrophobic interactions with receptor site.
Footnotes
Citation:Gnanavel & Muthukumar, Bioinformation 7(3): 112-114 (2011)
References
- 1.Niccolai, et al. Protein Sci. 2001;10:1498. doi: 10.1110/ps.30101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.T Tancredi, et al. Eur J Biochem. 2004;271:2231. doi: 10.1111/j.1432-1033.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 3.G Nelson, et al. Cell. 2001;106:381. [Google Scholar]
- 4.X Li, et al. Proc Natl Acad Sci U S A. 2002;99:469. [Google Scholar]
- 5.WB Floriano, et al. J Mol Model. 2006;12:931. doi: 10.1007/s00894-006-0102-6. [DOI] [PubMed] [Google Scholar]
- 6.Bassoli, et al. Journal of the Chemical Society Perkin Transactions. 1998;2:1449. [Google Scholar]
- 7.M Kohmura, et al. Pure & Appl Chem. 2002;74:1235. [Google Scholar]
- 8.DE Walters. Pure & Appl Chem. 2002;74:1117. doi: 10.1351/pac200274071135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.PA Temussi, et al. FEBS Letters. 2002;526:1. doi: 10.1016/s0014-5793(02)03155-1. [DOI] [PubMed] [Google Scholar]
- 10.SC Lovell, et al. Proteins. 2003;50:437. [Google Scholar]
- 11.D Duhovny, et al. WABI 2002 LNCS. 2002;2452:185. [Google Scholar]
- 12.D Schneidman-Duhovny, et al. Nucleic Acids Res. 2005;33:W363. [Google Scholar]