Figure 6.
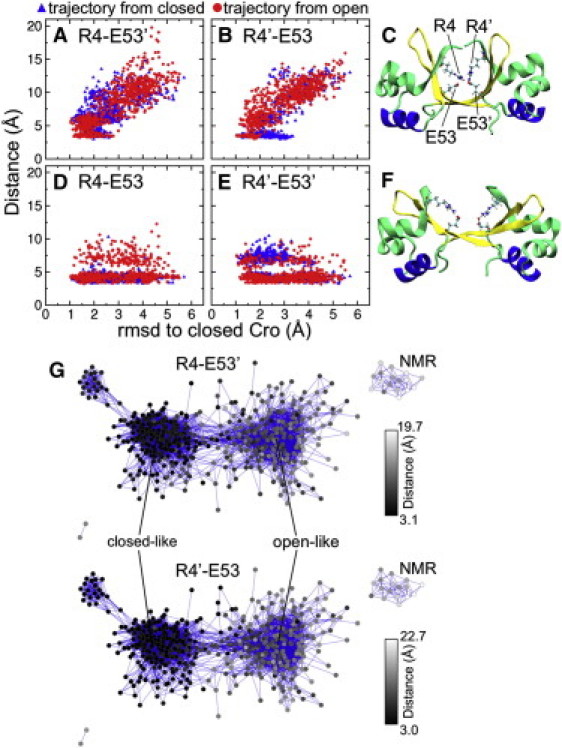
(A and B) Correlation between the intersubunit salt bridge distance (Arg4-Glu53′ and Arg4′-Glu53) and the RMSD to the closed crystal structure. (C) The structure from the combined REMD trajectory closest to the closed Cro x-ray image (1.0 Å RMSD) displays the intersubunit salt bridge between R4 and E53′. (D and E) Correlation between the intrasubunit salt bridge distance (Arg4-Glu53 and Arg4′-Glu53′) and the RMSD to closed Cro. (F) The conformation most like open Cro (1.0 Å RMSD) shows the intrasubunit salt bridges between these residues. (G) Intersubunit salt bridging as highlighted by the network layout. The Arg4-Glu53′ and Arg4′-Glu53 distances are represented as a color gradient in the network; black corresponds to shorter distances, and gray-white indicates larger distances. All data shown are for the last half of simulation (15–30 ns). Distances were measured between the centers-of-mass of Nη1 and Nη2 of arginine, and Oε1 and Oε2 of glutamate.
