Abstract
Horizontal gene transfer (HGT) is a common event in prokaryotic evolution. Therefore, it is very important to consider HGT in the study of molecular evolution of prokaryotes. This is true also for conducting computer simulations of their molecular phylogeny because HGT is known to be a serious disturbing factor for estimating their correct phylogeny. To the best of our knowledge, no existing computer program has generated a phylogenetic tree with HGT from an original phylogenetic tree. We developed a program called HGT-Gen that generates a phylogenetic tree with HGT on the basis of an original phylogenetic tree of a protein or gene. HGT-Gen converts an operational taxonomic unit or a clade from one place to another in a given phylogenetic tree. We have also devised an algorithm to compute the average length between any pair of branches in the tree. It defines and computes the relative evolutionary time to normalize evolutionary time for each lineage. The algorithm can generate an HGT between a pair of donor and acceptor lineages at the same evolutionary time. HGT-Gen is used with a sequence-generating program to evaluate the influence of HGT on the molecular phylogeny of prokaryotes in a computer simulation study.
Availability
The database is available for free at http://www.grl.shizuoka.ac.jp/˜thoriike/HGT-Gen.html
Background:
Since the whole genome of many species has been sequenced, methods have been developed to analyze all the available genes together evolutionarily in the genome of a species. One of the most popular methods is to construct a phylogenetic tree by using the data on all available of genes or proteins. Systematic evaluation of those methods is important. Perhaps, the only approach to this evaluation is computer simulation because it is impossible to conduct the evaluation using experiments or experimental data, particularly in the case of long-term evolution. Computer simulation programs such as ROSE [1], DAWG [2], SIMPROT [3], EvolveAGene [4], and iSG [5, 6], are useful for such evaluations. They are able to handle point mutations, insertions/deletions (indels), and the heterogeneity of evolutionary rates of genes. However, they do not include horizontal gene transfers (HGTs) in the evaluation.
HGT is the process in which an organism transfers a part of its genome to an unrelated organism. HGT genes are disturbing factors in the construction of a species' phylogenetic tree because they are produced through a biological mechanism that does not follow the authentic inheritance. Therefore, the influence of HGT on phylogenetic tree construction should be removed. In practice, however, such removal is currently impossible. A practical resort is to estimate the influence by conducting the computer simulation that considers HGT. We developed a program called HGT-Gen that converts a given tree constructed without the influence of HGT to one with it. HGT-Gen takes HGT into account and moves an operational taxonomic unit (OTU) or a clade from one place to another in a given tree. It can also generate HGT between a pair of lineages at any given evolutionary time.
Methodology:
Averaging of Branch lengths:
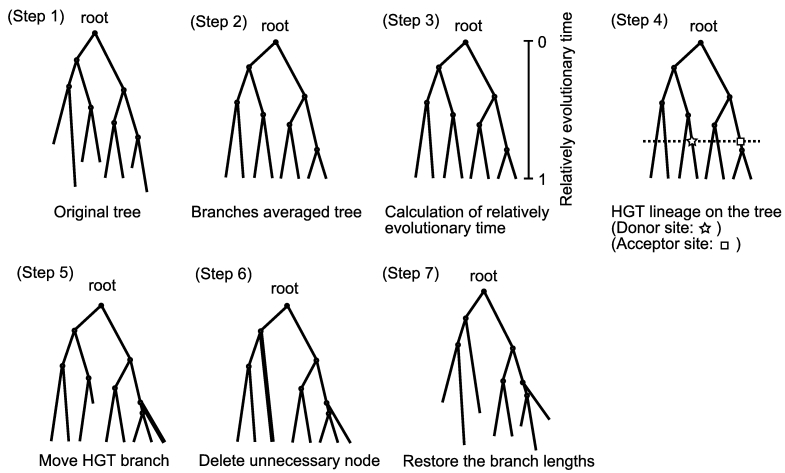
A rooted tree is needed as an input data that can be converted to a tree consisting of HGTs (Figure 1, Step 1). While the evolutionary times from the common ancestor to the present genes are the same, the corresponding branch lengths differ because of the stochastic nature of the mutations and other evolutionary factors. Therefore, we take the average length between every lineage pair (Figure 1, Step2). This process is recursively carried out from the bottom to the top of the tree.
Figure 1.
Algorithm of the HGT-Gen
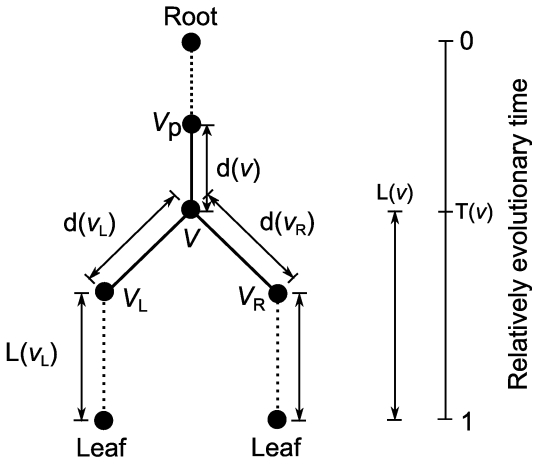
The averaged branch length from an arbitrary node v to each OTU is defined as follows: L(v) = (d(vL) + L(vL) + d(vR) + L(vR)) / 2, in which vL stands for the left descendant to v (Figure 2), vR stands for the right one, d(v) stands for the distance between v and vP, and vP stands for the parent of v. If v is an OTU, L(v) is 0.
Figure 2.
Scheme of the branch lengths from an arbitrary node v in a phylogenetic tree
Relative Evolutionary time:
To determine the evolutionary time of an HGT donor and its acceptor, the relative evolutionary time, T(v), for each lineage was computed (Figure 1, Step 3). T(v) on v is recursively determined using the following formula: T(v) = (d(v) + L(v)T(vp)) / (d(v) + L(v)), where if v is the root, T(v) is 0. On the other hand, if v is an OTU, T(v) is 1. This formula is derived on the condition in which {1 T(vp)}/{1 T(v)} = {d(v)+L(v)}/L(v).
Transfer of Branches:
A user first sets a time range and then randomly decides the time of an HGT event in the time range. The time range can be determined in the range between 0 (the root) and 1 (an OTU) in the normalized evolutionary time. An HGT donor lineage was randomly selected from the lineages located in the evolutionary time (Figure 1, Step 4). The HGT acceptor site of the selected lineage is randomly chosen from the lineages at the same evolutionary time. A new node is created correspondingly at the new location with the branch that is derived from the donor site (Figure 1, Step 5). The remaining branches at the donor site and the nearest upper node are then deleted (Figure 1, Step 6). The branch lengths are restored (Figure 1, Step 7).
Input:
HGT-Gen requires an input file containing the original phylogenetic tree in the Newick format. The time range (start and end) of the HGT events in relative evolutionary time and the number of the HGT trees in the output file are also required. An example of the command is as follows: perl hgtgen.pl input.tre 0.5 0.8 10 >output.tre In this case, the input file name is “input.tre”, the start position is 0.5, the end position, which must be equal or larger than the start position, is 0.8, the number of output trees is 10, and the output file name is “output.tre”.
Output:
The output file contains the phylogenetic trees with HGT in the multi-Newick format.
Utility:
HGT-Gen does not directly contribute to solving the HGT problems on phylogenetic analysis, but it provides artificial HGT data for studying precisely the influences of HTG on the phylogenetic methods such as those of constructing an ortholog dataset and a phylogenetic tree.
Future Development and Caveats:
The current version of HGT-Gen generates a single HGT. To generate more than one HGT on a phylogenetic tree, one must accordingly run HGT-Gen more than one time on the tree. We have a plan to develop the program to generate multiple HGT events. The execution speed is very fast. For example, in case of 10,000 trees with 64 OTUs each, the execution time is less than two minutes on a common PC. Therefore, HGT-Gen is almost limitless to the numbers of OTUs and output trees.
Acknowledgments
This work was supported by Grant-in-Aid for Young Scientists (B) No. 22710184.
Footnotes
Citation:Horiike et al, Bioinformation 7(5): 211-213 (2011)
References
- 1.RA Cartwright. Bioinformatics. 2005;21:iii31. [Google Scholar]
- 2.BG Hall. Mol Biol Evol. 2008;25:688. doi: 10.1093/molbev/msn008. [DOI] [PubMed] [Google Scholar]
- 3.A Pang, et al. BMC Bioinformatics. 2006;6:236. [Google Scholar]
- 4.J Stoye, et al. Bioinformatics. 1998;14:157. [Google Scholar]
- 5.CL Strope, et al. Mol Biol Evol. 2007;24:640. doi: 10.1093/molbev/msl195. [DOI] [PubMed] [Google Scholar]
- 6.CL Strope, et al. Mol Biol Evol. 2009;26:2581. doi: 10.1093/molbev/msp174. [DOI] [PMC free article] [PubMed] [Google Scholar]