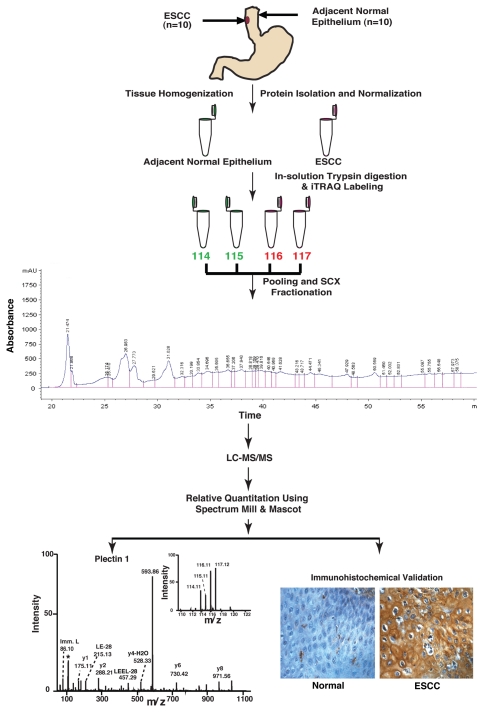
Figure 1.
Work flow for quantitative tissue proteomics using iTRAQ labeling and validation of biomarkers for esophageal squamous cell carcinoma. For iTRAQ labeling, proteins were isolated from ten tumor and adjacent normal tissues. Proteins were subjected to trypsin digestion followed by iTRAQ labeling of peptides. Post labeling of peptides the tumor and adjacent normal derived peptide mixture was pooled and fractionated using strong cation exchange (SCX) chromatography, followed by liquid chromatography tandem mass spectrometry (LC-MS/MS) on a QTOF mass spectrometer. The data was searched using Mascot and Spectrum Mill search engines. Some of the overexpressed proteins that were not previously described (e.g., PLEC1) were validated using IHC labeling using tissue microarrays.