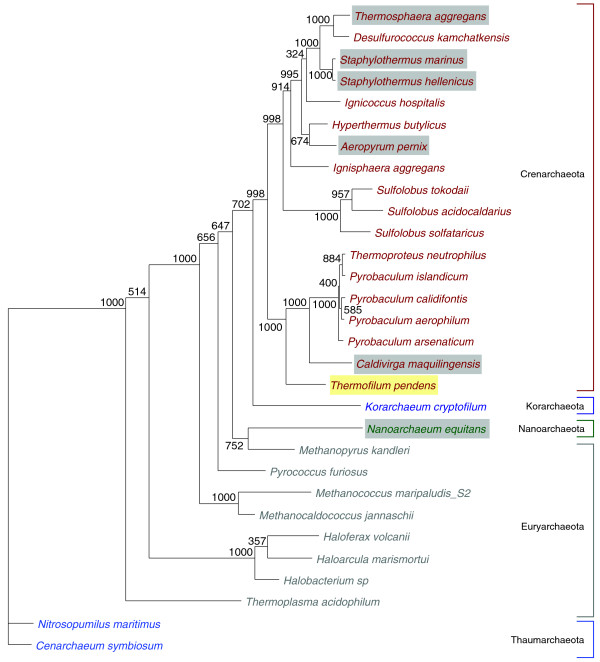
Figure 4.
Phylogenetic distribution of trans-spliced and permuted tRNAs in Archaea. Trans-spliced tRNAs were identified in A. pernix, T. aggregans, S. marinus, and S. marinus in this study, in N. equitans by Randau and colleagues [4,5], and in C. maquilingensis by Fujishima and colleagues [8] (highlighted in gray). Permuted tRNAs were found in T. pendens in this study (yellow), and are most similar to the permuted tRNAs found in Cyanidioschyzon merolae, a eukaryotic algal species that is not included in the tree but would be a distant outgroup to the archaea shown here. The phylogenetic tree was generated based on the concatenation of 23S and 16S ribosomal RNAs. Sequences were aligned using ClustalW [48]. Alignments were manually adjusted using Jalview [49] to remove introns. The maximum likelihood tree was computed using PhyML [50] with general time-reversible model of sequence evolution. Numbers at nodes represent non-parametric bootstrap values computed by PhyML [50] with 1,000 replications of the original dataset.