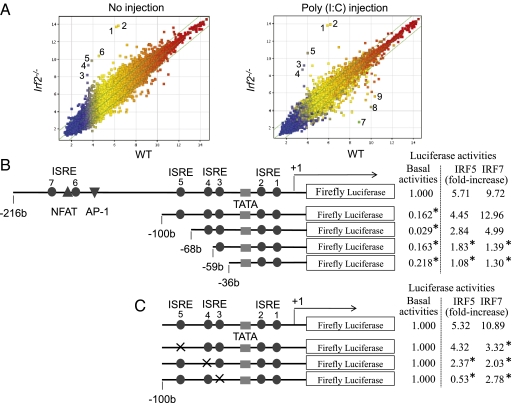
Fig. 2.
Trypsinogen5 is highly expressed in Irf2-deficent mice. (A) Irf2−/− or wild-type mice with or without peritoneal injection of pIC were killed, and the amounts of mRNA from the pancreas were systematically compared using Affymetrix 28,815 gene probes. The points farthest from the diagonal indicate transcripts showing the greatest difference between WT and Irf2−/−. Points 1 and 2, trypsinogen5 with different probes; 3, α-2-HS-glycoprotein (Ahsg); 4, annexin A10 (Anxa10); 5, fetuin-β (Fetub); 6, 3-hydroxy-3-methylglutaryl-CoenzymeA synthase2 (Hmgcs2, HMG-CoA synthase); 7, Ig κ chain variable8 (Igk-V8); 8, unknown; 9, carbonic anhydrase 3 (Car3). (B) A series of deletion mutants of trypsinogen5 proximal promoter region (−216 to +15) was placed upstream of a luciferase reporter gene (1 μg) and analyzed for transcriptional activity in mouse pancreatic acinar cells using a dual luciferase assay at 24 h posttransfection in combination with expression vectors (100 ng) expressing IRF5 or IRF7 or a control vector. The basal luciferase activity of each deletion, measured relative to the −216 to +15 region, and the responses to IRF5 and IRF7 expression vectors are shown as fold increase compared with the control vector. The TATA box, ISRE core, and NFAT- and AP-1 binding sites are indicated. *P < 0.05 versus the −216 to +15 region. (C) Point mutations were introduced into each ISRE site (indicated by x) of the trypsinogen5 promoters as described in Materials and Methods. The promoter activity of each mutant trypsinogen5 was determined with a dual luciferase assay system. *P < 0.05 versus wild type.