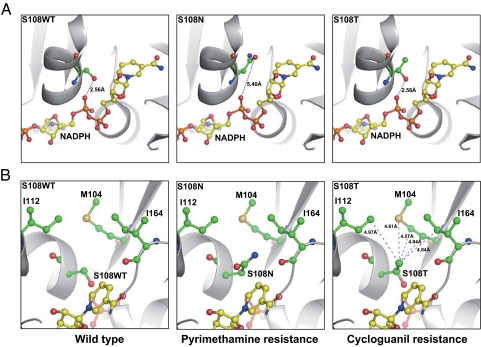
Fig. 4.
Key structural variances at amino acid 108 in WT and resistant mutants of P. falciparum DHFR. Represented are crystal structures of WT DHFR (S108WT) and S108N mutant (PDB ID codes 1J3I and 3JSU). The structure of the S108T mutant was derived by molecular modeling. (A) Note a 2.56-Å hydrogen bond between the pyrophosphate moiety of NADPH and the hydroxyl group of the S108 WT or S108T residue; no effective hydrogen bond exists in the S108N mutant due to the 5.40-Å distance. (B) Note extra methyl group in the S108T mutant forms van der Waals interactions with a nearby hydrophobic area composed of Met-104, Ile-112, and Ile-164. The side chains of S108WT, S108T, S108N, Met-104, Ile-112, Ile-164, and NADPH are highlighted with sticks and spheres; the hydrogen bond is shown as the dashed line with the distance labeled, and the protein backbone is shown in a cartoon model.