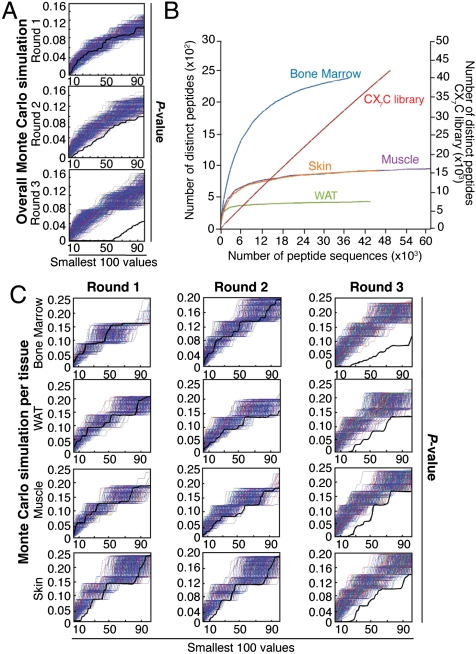
Fig. 1.
Combinatorial selection in patients. (A) Monte Carlo simulations with peptides analyzed in serial rounds of selection show nonrandom distribution of tripeptides. Thick black lines represent the Fisher’s exact test; thin blue lines represent the corresponding random permutated dataset. (B) A saturation plot (modified from ref. 17) shows the number of distinct peptides as a function of the total number of peptide sequences. All tissues reached saturation, as indicated by flattening of the slope; in contrast, unselected library showed no evidence of saturation. (C) Isolated peptides were grouped according to tissue-of-origin and subjected to Monte Carlo simulation. For every simulation, the pool of peptides was randomly distributed into groups corresponding to the number of sequences analyzed for each targeted tissue. Frequencies were calculated for each simulated organ, and Fisher’s exact test applied on the permutated dataset. A nonrandom selection of tripeptides was observed in all organs tested.