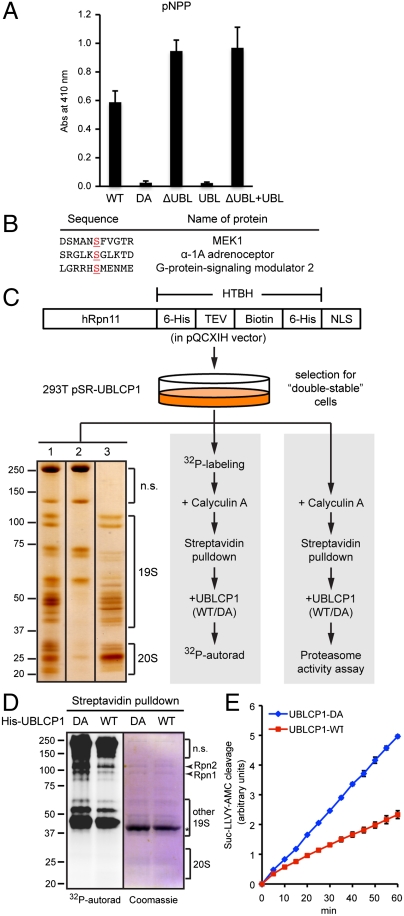
Fig. 3.
UBLCP1 dephosphorylates the proteasome in vitro. (A) pNPP assay with the indicated UBLCP1 proteins. Data are shown as average ± SD. (B) Phospho-peptides significantly dephosphorylated by UBLCP1 in vitro, with the phospho-serine residues underlined. This phospho-peptide array (Millipore) does not include any proteasome peptides. (C) 26S proteasome isolation and analysis. 293T double-stable line was used for high-purity proteasome preparation (Left), 32P-labeling and in vitro phosphatase assay (Middle), or in vitro proteasome activity assay (Right). (Left) A representative silver-staining image of streptavidin-immobilized proteasomes (lane 1), purified proteasomes after tobacco etch virus (TEV) protease treatment (lane 3), and nonspecific (n.s.) proteins remaining on the beads (lane 2). See Materials and Methods for details. (D) Metabolically radio-labeled, affinity-purified nuclear proteasomes were treated in vitro with purified UBLCP1 (shown by an asterisk). Proteasome subunits were visualized by Coomassie blue staining and 32P-autoradiography. (E) Peptidase activity assay of the 26S proteasome purified and treated as in D. AMC fluorescence is represented as the average ± SD.