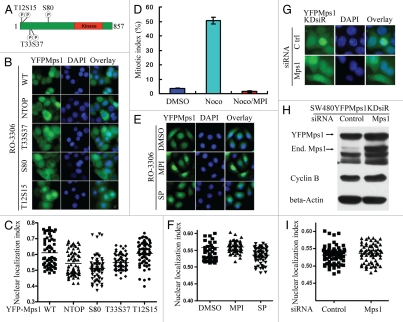
Figure 3.
The kinase activity of Mps1 is not required for Mps1 nuclear entry. (A) The relevant phosphorylation sites in the N terminus of Mps1 are shown. (B) Subcellular localization YFP-tagged Mps1 and Mps1 mutants in SW480 cells were imaged after RO-3306 treatment for 19 h. Shown were representative images for each mutant. At least 50–70 cells were inspected. (C) A scatter plot of the distribution of results obtained in (B). The average distribution for each sample is shown as a horizontal bar. Using SNK grouping and the Kruskal-Wallis test, the difference of nuclear localization index between YFPMps1T33S37AA and NTOP is not identified as significant (p > 0.05), while the difference between S80 and NTOP is significant (p < 0.0001). There is no statistical difference between WT and T12S15 (p > 0.05). (D) MPI-0479605(MPI), a specific inhibitor of Mps1 kinase activity abolishes the spindle assembly checkpoint arrest induced by nocodazole (100 ng/ml). (E) Nuclear accumulation of YFP-Mps1 is unaffected by co-treatment with MPI-0479605 (10 µM or SP600125 (10 µM) and RO-3306 for 12 h after released from double thymidine treatment. (F) A scatter plot of the distribution of YFP-Mps1 treated with control, MPI-0479605 or SP600125. About 50–70 cells for each treatment were counted and quantified. (G) Subcellular localization of YFP-Mps1KD upon treatment with RO-3306 in the presence or absence of the endogenous Mps1. SW480 cells stably expressing YFP-Mps1KDsiR were transfected with either control or Mps1 siRNA. (H) Depletion of the endogenous Mps1 is verified by immunoblotting with an anti-Mps1 antibody (Millipore). β-actin was blotted as loading control. Cyclin B expression is indicative of G2/M arrest. (I) Quantitation plot of subcellular localization of YFP-Mps1KDsiR treated with control or Mps1 siRNA. There is no significant statistical difference among DMSO, SP and MPI compound groups or between control and Mps1 siRNA groups (p > 0.05) in (F and I).