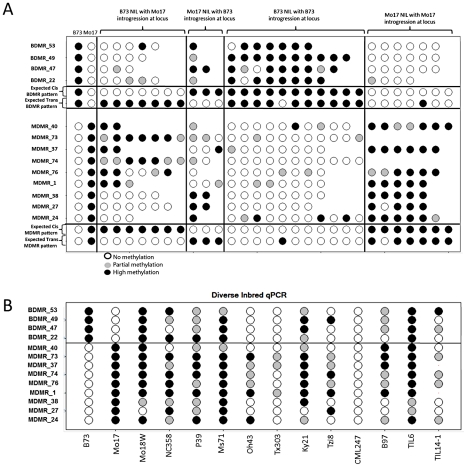
Figure 4. Variable DNA methylation patterns in near-isogenic lines and diverse inbreds.
(A) The relative DNA methylation levels in selected near-isogenic lines was tested by digestion with the methylation dependent restriction enzyme MspjI followed by qPCR. Different subsets of NILs were selected and analyzed for each of 13 DMRs. The first two columns show the data from B73 and Mo17. Open circles reflect low methylation levels and black circles indicate high methylation levels. Intermediate methylation levels are indicated by gray color. The next group of 2–7 genotypes show the data from NILs that have B73 as the recurrent parent (>95% of the genome) and have introgression of the Mo17 haplotype in the region containing the DMR. The variable number of genotypes tested reflects the fact that some DMR loci are have more NILs with an introgression than others. The next group of 1–3 genotypes are NILs that are primarily Mo17 but have B73 introgressed at the DMR. The next two groups provide “control” genotypes of B73-like or Mo17-like NILs that do not have an introgression at the DMR. The expected patterns for cis (local) inheritance of DNA methylation or trans (unlinked) control of DNA methylation are shown. Note that the expected pattern for trans control would include a small number of genotypes with the methylation pattern from the introgressed genotype in cases where the trans-acting locus is introgressed. (B) The same type of assays were performed on a panel of 12 diverse inbred genotypes, including two inbred teosinte lines, to monitor the frequency for the hyper- and hypo-methylated states.