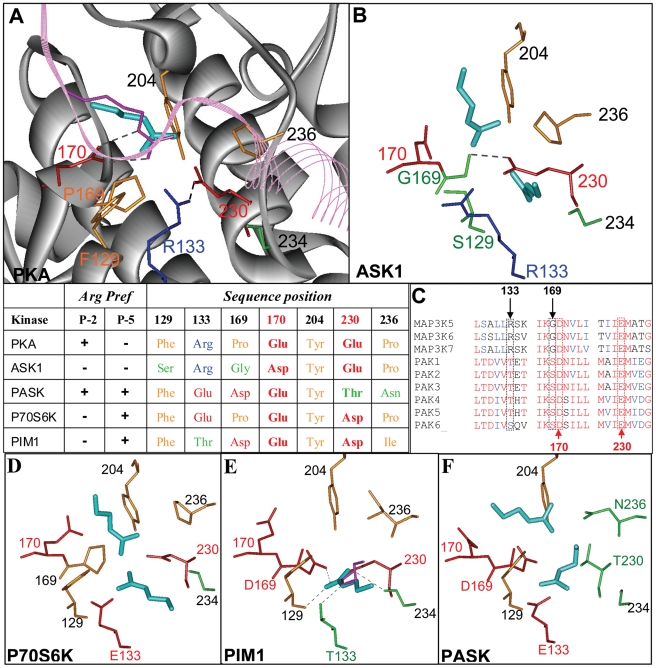
Figure 6. Key positions composing the −2/5 site of several kinases.
Residues at positions: 129, 133, 169, 170, 204, 230, 236 and the backbone of position 234 are presented. For clarity, hydrophobic amino acids: Tyr, Trp, Ile and Pro are colored orange. Carbonyl and polar uncharged amino acids: Thr, Ser, Asn and also Gly are colored green. Negatively charged (Asp, Glu) and positively charged (Arg) amino acids are colored red and blue, respectively. Experimentally deduced H-bonds are shown as dotted gray lines. Predicted (panels A,B,D,E and F) and experimental (panels A and D) Arg-binding positions are colored in cyan and magenta respectively. For kinases for which an experimental Arg-binding position exists, the presented solution is of the lowest RMSD; otherwise, the lowest energy prediction in each subsite is presented. The table summarizes the key residue contents of all five kinases. (A) Viewing key residues at the −2/5 site of the PKA/PKI complex (1ATP). The PKI peptide is colored in cyan. (B) The predicted Arg positions and key residues at the −2/5 sites of ASK1. (C) Sequence alignment of 9 kinases from the STE family with an acidic-pair pattern. (D–F) Key residues and predicted Arg-binding positions at the −2/5 sites of P70S6K, PIM1 and PASK, respectively. For PIM1, the experimental Arg-binding position is shown in magenta.