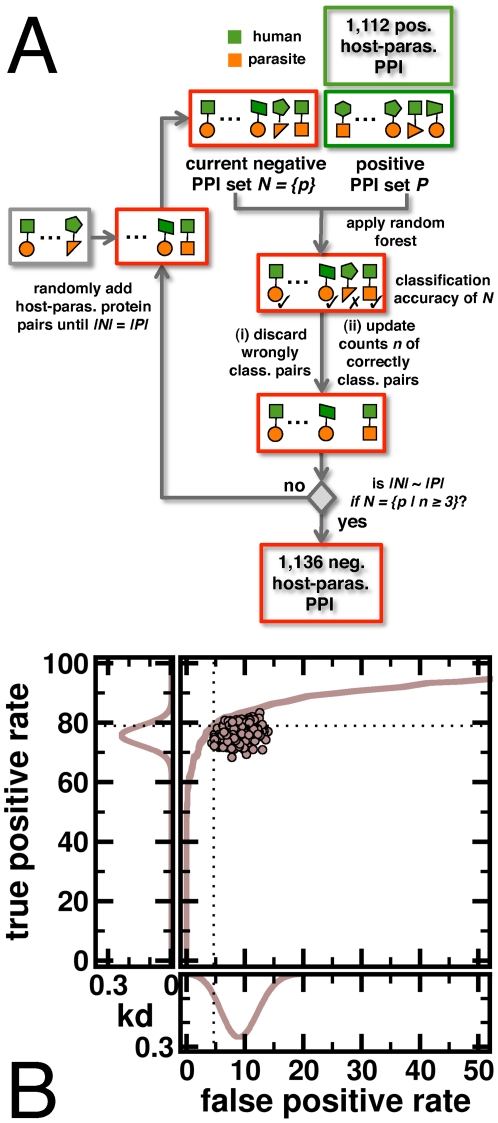
Figure 2. Determination of a negative interactions set.
(A) As a positive training set I utilized 1,112 interactions between the human host and the parasite P. falciparum that have been previously inferred from protein structures. A non-interacting training set of equal size was constructed by randomly sampling pairs of human and parasite proteins that did not appear in the positive training set. Applying the random forest algorithm pairs of proteins that were incorrectly classified as interacting were discarded, and counts of correctly classified pairs were updated. If the number of pairs in the negative set that were sampled at least 3 times was roughly the size of the positive training set, the procedure terminated. Otherwise, the negative set of was filled with randomly sampled protein pairs until positive and negative training sets had the same size again. Previously described steps were repeated until the procedure finally terminated, providing a negative set of 1,136 non-interacting pairs. (B) Applying the random forest algorithm the training sets allowed for a true positive rate TPR = 78.9% and a false positive rate FPR = 4.7% (dashed lines) in a pronounced ROC curve. In a test-retest analysis, the classifier was trained on ⅔rd of randomly picked training data. Its performance was tested on the remaining ⅓rd, indicating that the proposed method was largely robust to noise.