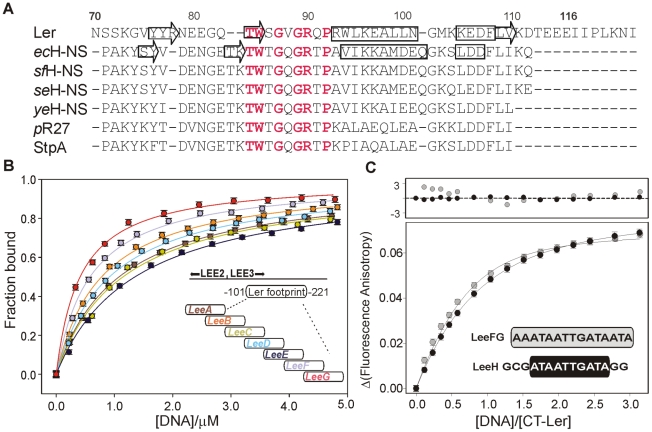
Figure 1. DNA-binding domain of selected members of the H-NS family of proteins and DNA fragment optimization.
(A) Sequence alignment of the C-terminal domain of the following proteins: Ler; chromosomal H-NS of E. coli (ecHNS); Shigella flexneri (sfHNS); Salmonella enterica serovar Typhimurium (seHNS); Yersinia enterocolitica (yeHNS); the plasmid R27-encoded H-NS protein (pR27); and E. coli StpA. The secondary structure elements of DNA-bound CT-Ler and free H-NS are shown. Highly conserved residues within the consensus DNA-binding motif are highlighted in red. (B) Analysis of the interaction of CT-Ler with 30 bp DNA fragments (LeeA-G, sequences are listed in Table S1) derived from the DNAse I footprint of Ler in the LEE2/LEE3 regulatory region [10]. Complex formation was followed by the increase of CT-Ler fluorescence anisotropy. (C) Fluorescence anisotropy titrations of CT-Ler with LeeH (black circle) and LeeFG (gray circle). Solid curves are the best fit to a model assuming a 1∶1 complex. The point by point deviations between fitting and experimental points are shown in the top panel.