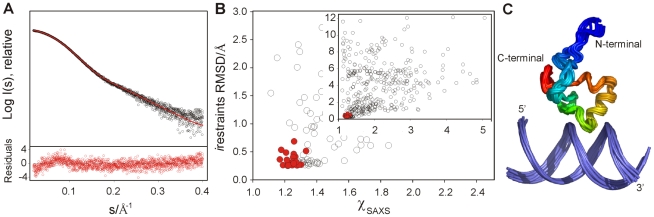
Figure 3. Structure determination of the CT-Ler/LeeH complex based on NMR and SAXS.
(A) SAXS intensity in logarithmic scale measured for a CT-Ler/LeeH equimolar sample (open circles) as a function of the momentum transfer 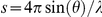 , where
, where 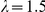 Å is the X-ray wavelength and
Å is the X-ray wavelength and 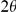 is the scattering angle. CRYSOL fit of the SAXS curve using a representative NMR structure (red); the average deviation
is the scattering angle. CRYSOL fit of the SAXS curve using a representative NMR structure (red); the average deviation 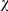 is 1.16. Only the range 0.018< s <0.4 Å−1 is displayed. The point by point deviations [(I(s)exp−I(s)fit)/
is 1.16. Only the range 0.018< s <0.4 Å−1 is displayed. The point by point deviations [(I(s)exp−I(s)fit)/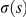 ], where
], where 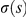 is the experimental error are shown in the bottom panel. (B) Scatter plot of NMR intermolecular restraint violations versus
is the experimental error are shown in the bottom panel. (B) Scatter plot of NMR intermolecular restraint violations versus 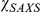 values for the initial set of 400 complex structures and the final ensemble of 20 low energy structures highlighted in red (inset). The main panel shows a zoom of the best structures. (C) Backbone overlap of the 20 lowest energy complex structures. Protein backbone is coloured in rainbow gradation.
values for the initial set of 400 complex structures and the final ensemble of 20 low energy structures highlighted in red (inset). The main panel shows a zoom of the best structures. (C) Backbone overlap of the 20 lowest energy complex structures. Protein backbone is coloured in rainbow gradation.