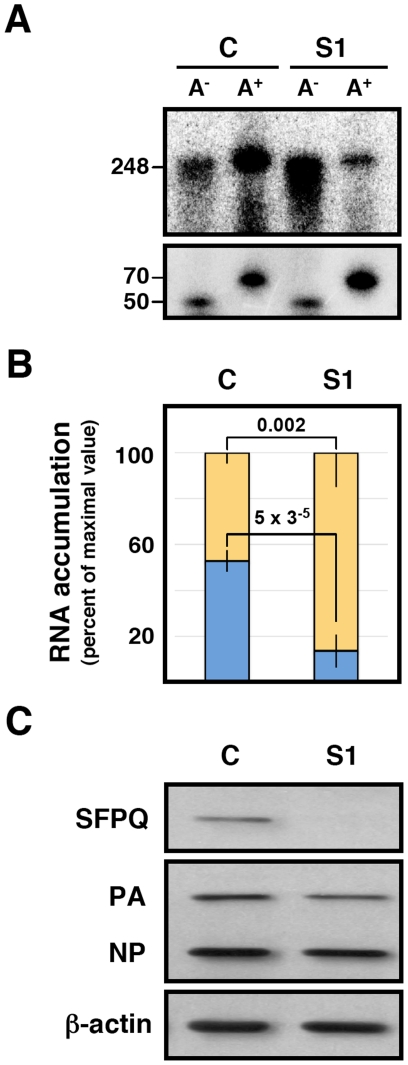
Figure 12. Dependence of SFPQ/PSF for the in vitro polyadenylation of transcripts.
Recombinant RNPs were generated in control-silenced (C) or SFPQ/PSF-silenced (S1) HEK293T cells and in vitro transcription was performed as indicated in the legend to Figure 11. The transcripts were separated into poly A+ and poly A− fractions, using a non-polyadenylated oligonucleotide (50 nt) and a polyadenylated oligonucleotide (70 nt) as recovery probes, and the fractionated transcripts were analysed by electrophoresis on denaturing polyacrylamide gels. (A) Representative results of five independent experiments. The positions of a 248 nt marker identical to the clone 23 genome, as well as the recovery probes are indicated to the left. (B) Quantification of the proportion of the poly A+ (blue) and poly A− (yellow) transcripts after normalisation for recovery. The data represent averages and standard deviations of five experiments. Significance was determined by the Student's t test. (C) The silencing of SFPQ was controlled by Western-blot, using actin as loading control. The level of expression of PA and NP was ascertained by Western-blot.