Abstract
Background:
Early childhood caries (ECC) is a common disease process that afflicts a large proportion of the child population worldwide. Extensive research in past indicates that it is the result of bacterial infection, also influenced by host and dietary factors. Current caries research seeks to identify risk factors as well as natural oral defenses that may protect against or prevent caries development. Saliva, in spite of being the strongest defense system, still has a wide array of properties and proteins whose role is yet not clearly known.
Aim:
To compare the resting human whole salivary characteristics in children with ECC and those who are caries free. Settings and Design: The study was conducted over a period of 9 months in 4- to 6-year-old 100 children comprising two groups – 50 with ECC and 50 caries free.
Materials and Methods:
The whole salivary flow rate, pH, mean protein concentration, and the electrophoretic profile of salivary proteins by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) were compared among both groups.
Statistical Analysis:
The SPSS (version 11.0) software package was used to conduct the chi-square, Fisher's exact and Pearson's chi-square tests to compare the data.
Results:
On gel electrophoresis, there was a significant difference among both groups with caries-free subjects having a higher number of proline-rich protein bands, substantiating the protective role of this protein. A significantly higher number of glycoprotein bands were observed in the whole saliva of subjects with ECC. A significant inverse correlation between the mean protein concentration and the whole salivary flow rate was observed in both groups.
Keywords: Amylase, early childhood caries, gel electrophoresis, mucins, proline-rich proteins
Introduction
Saliva is one of the most important protective body fluids but it still remains the least understood. Though numerous studies in the past have attempted to relate certain aspects of salivary output and composition to caries susceptibility in children, however, predominantly these studies looked at either the physiochemical properties of saliva (flow rate, buffer capacity) or specific components of saliva with an antimicrobial activity, such as salivary IgA, lactoferrin, lysozyme, and the salivary peroxidase–hypothiocyanate system. With the possible exception of the hypothiocyanate system and IgA, the earlier consensus was that a relationship between caries experience and an activity of any of the salivary antimicrobial proteins could not be demonstrated in children.[1]
In spite of advances in correlating various risk factors to caries in children, and newer treatment modalities and preventive strategies, a large fraction of the world-wide population of children is still affected significantly by “early childhood caries” (ECC).
Variabilities in salivary proteins and their posttranslational modifications may play an important role in determining their protective features toward dental caries. It has thus been agreed that genetic factors associated with the phenotypic expression of these proteins in mixed saliva can contribute to the evaluation of dental caries risk in children and hence early prevention of this widespread disease. Thus, the exploration of these inherited markers for ECC is very important.[2]
Recently, there has been a growing interest in exploring the protein composition using advanced proteomics technology. Proteomics is a powerful approach for biomedical research because it directly studies the key functional components of biochemical systems and the cellular targets of therapeutics agents, namely proteins. In a typical proteomic approach, the separation of proteins and their visualization using a stain is the key process. Gel electrophoresis is one of the important tools now available to separate proteins based on their relative mobility on application of electric current depending on the molecular mass for evaluation.[3]
However, in spite of these technological advances in the field of proteomic research, not many studies have attempted to decode the array of salivary proteins particularly in children in order to determine the role of these proteins in ECC. The present clinical study was thereby undertaken to have a comparative evaluation of the electrophoretic profile of the salivary proteins in whole saliva of children suffering with ECC and those free of ECC by carrying out sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS–PAGE). In the case of specific characteristics and individual variability in the protein concentration and electrophoretic profile in the population studied, this study can provide the initial step for using the presence of types of salivary proteins as a risk predictor for ECC in children.
Materials and Methods
This study was conducted for a period of 9 months in the Department of Pedodontics, Manipal College of Dental Sciences, in collaboration with the Department of Biotechnology, Kasturba Medical College, Manipal, with an aim to have a comparative evaluation among caries-free children and children with ECC from the age group of 4–6 years in terms of the resting human whole salivary pH, flow rate, protein composition, and protein pattern on gel electrophoresis.
A total of 100 subjects were divided equally into two groups:
Group I – children with ECC
Group II – caries-free children.
Inclusion criteria
The criteria were as follows:
Subjects free from any systemic illness and not on any medication since past 1 month.
Subjects in Group I – those with a dmft/DMFT score (as per the WHO diagnostic criteria, 1986[4]) of >6, with carious involvement of maxillary incisors and at least two posterior teeth in either maxillary or mandibular arches.
Subjects in Group II – those absolutely free from caries with a dmft/DMFT score (as per the WHO diagnostic criteria, 1986[4]) of 0.
Protocol approval was obtained from Kasturba Hospital Ethics Committee, Manipal. Before the commencement of the study, the parents were clearly explained the purpose of the study and informed consent was obtained from them.
Clinical examination of all the subjects was carried out by a single examiner. The caries status for the subjects in Group I was determined with decayed, missing, or filled teeth (DMFT/dmft) index and calibrated according to World Health Organization (1986)[4] diagnostic criteria. All the subjects selected for Group II were absolutely caries free. To rule out the intra-examiner variability, replicate readings were taken randomly for 20 subjects. The subjects were explained the purpose and the method of collection of saliva, which was not done on the day of clinical examination.
Resting human whole saliva of all subjects was collected in a quiet room in the morning time as per the method suggested by FDI.[5] The salivary samples were labeled according to the group to which the subject belonged (numerically 1, 2, 3…. for Group I and alphabetically A, B, C…..for Group II).
Subjects were instructed to accumulate saliva in the mouth for 2 min and then spit it in the receiving vessel, which was a sterile plastic bottle. The same procedure was repeated three times. Samples were then transferred to a calibrated cylindrical flask which was used to measure the volume of saliva collected for each subject. The salivary flow rate was calculated as follows:[5]
Flow rate (mL/min) = Total volume of saliva collected divided by 6. The value obtained was noted.
A drop of each subject's saliva was put on the Dental Saliva pH indicator strips (pH 5.0–8.0, GC Corp., Japan) and the pH value was noted based on the color change of the strip when compared to the color coded chart.[5]
The saliva samples were transferred to 2.0-mL microcentrifuge tubes (Tarsons, Calcutta, India) which were labeled and samples were clarified by centrifugation using Spinwin MC-01 microcentrifuge (Tarsons, Calcutta, India) at 10,000 rpm for 5 min. The supernatant was then transferred to another microcentrifuge tube and these samples were stored at –80°C (–80°C refrigerator, Nuaire, USA) for further processing.
The samples were brought to room temperature and then centrifuged at 10,000 rpm for 5 min prior to dilution. The protein estimation of all the samples was done according to the spectrophotometric method suggested by Layne.[6]
The protein concentration of each sample was calculated in mg/mL using the following formula:
Protein concentration (mg/mL) = (1.56 × OD280 – 0.76 × OD260) dilution factor,
where the dilution factor for each sample was 20. Remaining saliva samples were frozen and stored at –80°C for gel electrophoresis and subsequent analysis.
Protein precipitation was done using ice-cold acetone and trichloroacetic acid. After this, samples were subjected to SDS–PAGE.
Prepared samples were analyzed by SDS–PAGE using the 12% resolving gel and 5% stacking gel. SDS–PAGE was carried out according to the method used by Schwartz et al.[7] with slight modifications. Ten samples (five from each group) were loaded in one gel. Two gels were run for all the samples at one time (one was stained using silver nitrate followed by Coomassie blue; the second gel was stained by α-naphthol). Molecular weight standards (Bio-Rad, California, USA) were used in all the gels. Electrophoresis was done at 100 V for around 3–4 h and then it was removed carefully for staining using the various reagents. Salivary proteins were identified according to their relative mobility in gel and stain patterns following the criteria described by Banderas-Tarabay et al.[2] Number of bands present for each subject was counted on the Coomassie-stained gel. Proline-rich protein (PRP-1) and amylase bands were scored according to the band size and stain intensity as absent (–), present (±), and high intensity and size (+) on Coomassie- [Figure 1] and silver-stained [Figure 2] gels. The glycoprotein bands on α-naphthol-stained [Figure 3] gels were scored as absent (–) or present (+).
Figure 1.
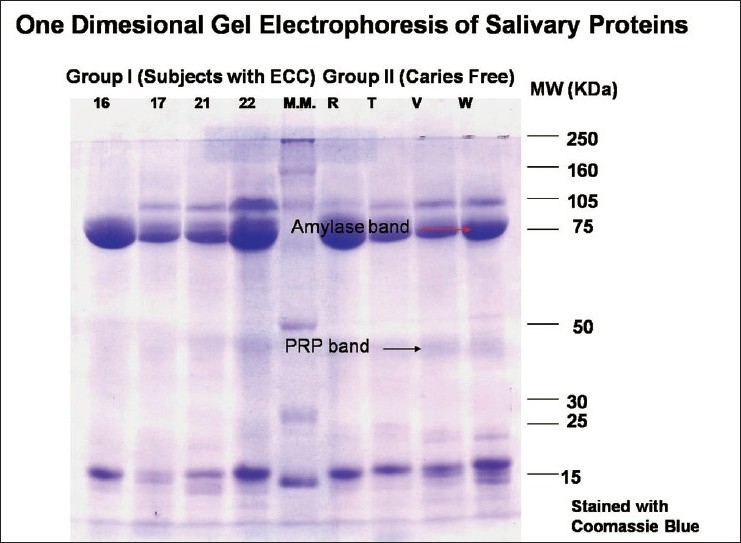
Coomasie-stained gel showing PRP and amylase bands
Figure 2.
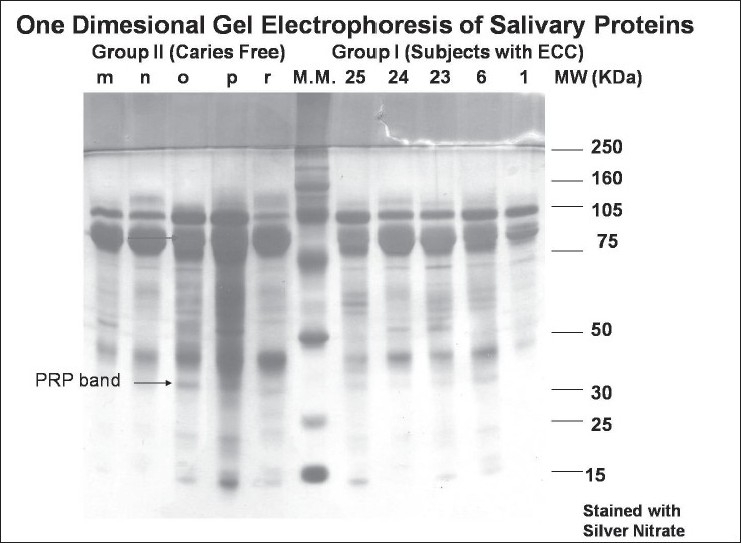
Silver nitrate-stained gel showing amylase and PRP bands
Figure 3.
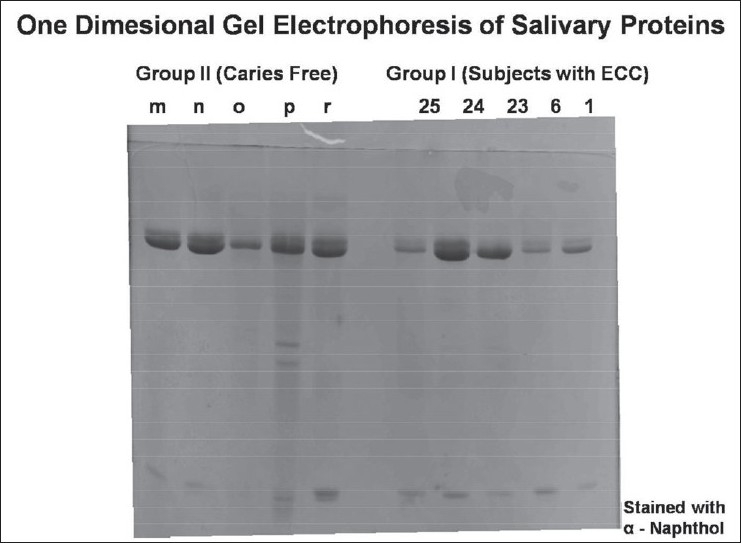
Alpha naphthol-stained gel for glycoproteins
Statistical analysis
All the data was entered into the SPSS (version 11.0) software package. The intergroup comparisons were carried out using the paired t-test, chi-square test, Fisher's exact test, and Pearson's chi-square test. The correlation between the pH, flow rate, and protein concentration was determined using Pearson's correlation coefficient.
Results
Mean whole salivary pH, flow rate, and protein concentration
The mean whole saliva pH, flow rate, and protein concentration values for Groups I and II were found to be statistically insignificant (P - value > 0.05).
Inter-group comparison of proline-rich protein bands [Table 1]
Table 1.
Inter-group comparison of proline-rich protein bands (using the chi-square test)
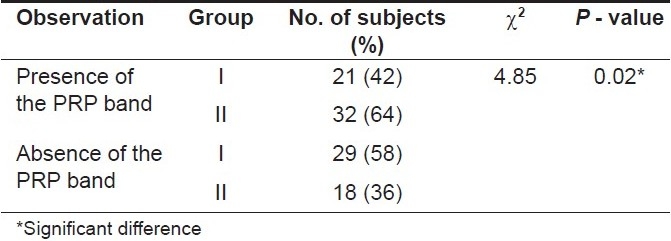
Proline-rich protein bands were observed in a total of 53 (53%) subjects. These bands were observed in 21 (42%) subjects in Group I whereas in Group II, these bands were observed in 32 (64%) subjects. The chi-square test was carried out. The P-value obtained on comparing both the groups was 0.02 (< 0.05) indicating that this difference was statistically significant suggesting that the caries-free subjects in Group II had a significantly higher number of proline-rich protein bands as compared to subjects with caries in Group I.
Inter-group comparison of amylase bands [Table 2]
Table 2.
Inter-group comparison of amylase bands (using the Fisher's exact test)
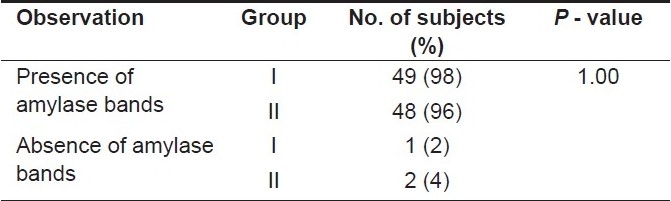
Amylase bands were observed in a total of 97 (97%) subjects. These bands were observed in 49 (98%) subjects in Group I whereas in Group II these bands were observed in 48 (96%) subjects. Fisher's exact test was carried out to compare the amylase bands present in both the groups. The P-value obtained on comparing both the groups was 1.00 (>0.05) indicating that this difference was statistically insignificant.
Inter-group comparison of glycoprotein bands [Table 3]
Table 3.
Inter-group comparison of glycoprotein bands (using the chi-square test)
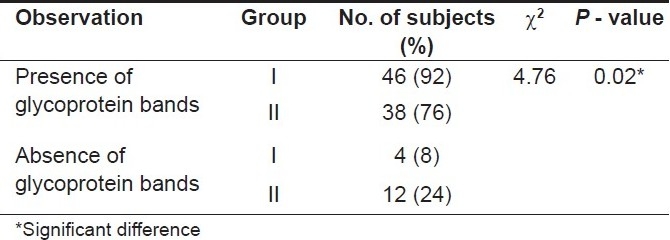
Glycoprotein bands were observed in 84 (84%) subjects. The bands on glycoprotein staining were observed in 46 (92%) subjects in Group I whereas in Group II these bands were observed in 38 (76%) subjects. The chi-square test was applied to compare the glycoprotein bands present in both the groups. The P-value obtained on comparing both the groups was 0.02 (< 0.05) indicating that this difference was statistically significant.
Inter-group correlation between the mean protein concentration and pH of whole saliva [Table 4]
Table 4.
Inter-group correlation between the mean salivary pH, flow rate, and protein concentration (using Pearson's correlation coefficient test)
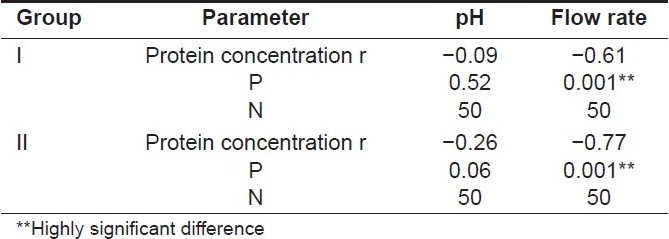
An increase in the whole saliva pH was observed in both the groups with the decreased protein concentration, and Pearson's correlation coefficient test was applied to establish the correlation among both variables. The P-values obtained were 0.52 for Group I and 0.06 for Group II which were >0.05 and hence this correlation was found to be statistically insignificant for both the groups.
Inter-group correlation between the mean protein concentration and the flow rate of whole saliva [Table 4]
An increase in the whole saliva flow rate was observed in both the groups with a decreased protein concentration and Pearson's correlation coefficient test was carried out to establish the correlation among both variables. The P -value obtained was 0.001 for both Groups I and II which was < 0.05 and hence this correlation was statistically highly significant for both the groups indicating that with an increased flow rate the mean protein concentration showed a highly significant decrease in all the subjects.
Discussion
Salivary defense systems including the salivary proteins play a significant role in maintaining the health of the oral cavity and preventing caries as has been agreed by Mazengoet al.[8]
The prediction of caries risk has been of long-standing interest and is very important for the development of new preventive strategies for caries. This is especially significant for young children. It has been suggested earlier that whole saliva is an easily available fluid which can be collected noninvasively and used to measure and monitor the risk for caries.[9] It has also been previously concluded by Dodds et al.[1] that the unstimulated state is the predominant condition in terms of the salivary gland activity, and the unstimulated saliva flow is the critical determinant of salivary clearance. Thereby unstimulated whole saliva of all the subjects was collected for analysis in this trial.
The difference in the mean whole salivary flow rate of both groups was statistically insignificant (P > 0.05) indicating that the inverse relationship between the whole salivary flow rate and the occurrence of dental caries in children as reported by Johansson et al.[10] was not observed in this study.
Similar results showing no association of parotid saliva flow rates with the occurrence of dental caries among either men or women or caries-active/caries-free groups have been reported earlier by Dodds et al.[1] This finding has been explained by Leone and Oppenheim[11] in their review on physical and chemical aspects of saliva as risk indicators of dental caries in humans. According to them, this negative result can be attributed to a number of confounding experimental factors. In particular, whenever differences in disease severity are minimal among groups, then it is very difficult to establish any effect on the caries status due to salivary flow.
Though a higher mean pH value of 6.61 ± 0.37 was obtained for caries-free subjects in Group II as compared to 6.48 ± 0.39 for the subjects with caries in Group I, this difference was not found to be statistically significant in the present trial. Colorimetric salivary pH determinations and the incidence of dental caries of 351 young male adults were studied for the first time by Carlton[12] but no relationship between the incidence of dental caries and the pH of normal resting saliva was evident in their work similar to this study.
In our study, the paired t-test was carried out to determine the level of significance between the mean protein concentrations of the two groups. The P-value obtained was 0.54 (>0.05) indicating that this difference however was not statistically significant. Similar to the present study, De Farias et al.[13] had evaluated the organic composition of saliva from children without dental caries and children with ECC. In their study, there were two groups with 20 subjects each and they also reported no significant difference in the mean protein concentration of saliva among both the groups.
An increase in the whole salivary flow rate was observed among both the groups with a decreased protein concentration suggesting that with an increased flow rate the mean protein concentration showed a highly significant decrease indicating an inverse relationship between the salivary flow rate and protein concentration. This finding was consistent with the observations by Cohen et al.[14] who have explained the increase in salivary flow rates resulting in decreased protein concentrations possibly due to protein dilution levels in saliva.
Though it can be seen that other parameters such as flow rate, pH, and mean protein concentration of whole saliva did not show any significant correlation with the caries status of the subjects in the study, nevertheless, without ignoring other possibilities, the main goal of the trial was to analyze the hypothetical relevance of the individual saliva composition to the susceptibility for the development of an oral infectious pathology such as ECC and hence the individual protein profile of each sample was analyzed depending upon the relative mobility of proteins on gel electrophoresis using the criteria specified by Banderas-Tarabay et al.[2]
Electrophoretic separation revealed a considerable variation in patterns of different individuals as shown by substantial differences in number, intensity, and size of observed bands. In this study, the presence of genetic polymorphism in salivary proteins of human whole saliva can thus be appreciated. The variability in protein profiles among subjects of this study is similar to that reported by Schwartz et al.[7] and Banderas-Tarabay et al.[2]
The presence of proline-rich protein bands was observed in a total of 53 (53%) subjects. This percentage was lower than 95% subjects having PRP bands in the population studied by Banderas-Tarabay et al.[2] However, this difference can be substantiated because it is well known that whole saliva contains fewer of the pink staining PRP bands than parotid and submandibular/sublingual saliva as suggested by Beeley.[15] It has been explained by the same author that the liability of the pink-violet staining PRP bands is a reflection of the variable loss of these proteins as they pass from the glands to the mouth. However, despite the reduction from the glandular PRP concentration, there remains significant “PRP-like” activity in whole saliva as the phosphopeptide fragments of acidic PRPs are present in whole saliva. When isolated, these fragments show biologically significant caries-protective effects. It is likely that many of the small bands in the present study gels represent these peptide fragments.
Similar to our results, Tenovuo[16] observed that acidic PRPs were significantly correlated with lower DMFT scores. The presence of acidic PRPs in higher relative abundances in the caries-free group emphasizes their protective role, especially considering their known role as enamel pellicle precursors and their involvement in the remineralization processes, which is in agreement with results reported by Banderas-Tarabay et al.[2]
The results of the present study are also consistent with the work done by De Farias et al.[13] In their results, children with ECC presented significantly higher levels of total salivary IgA and IgG, while the mean values of the amylase activity, total protein concentrations, and total IgM were similar between the groups. In the present trial also, there was no statistical significance between both the groups in terms of the presence of amylase bands.
In the present study, a significant difference was seen in the presence of glycoprotein bands on a-naphthol staining. More number of glycoprotein bands were observed in Group I subjects. Salivary mucins are known to have a protective role toward oral surfaces and their absence has been associated with an increased prevalence of dental caries in adults by Banderas-Tarabay et al.[2] whereas in the present study no such correlation was observed.
Many salivary protein components, such as proline-rich glycoproteins, mucins, immunoglobulins, agglutinin, lactoferrin, cystatins, and lysozyme, are thought to have a role in defense in the oral cavity.[17] Numerous studies have investigated the correlation between these salivary proteins and glycoproteins and caries experience, but no studies have shown a reliable association between a single salivary component and the caries experience.[18–19] In the present study also a significant correlation of the presence of PRP bands in caries-free subjects of Group II was the only positive finding.
Though the results are not highly conclusive as ECC is a multifactorial disease and thus linking it to only one risk factor is not possible, however further information on the molecular epidemiology of salivary proteins can support use of this methodology as a diagnostic tool in ECC and other oral health problems in young children.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Dodds MW, Johnson DA, Mobley CC, Hattaway KM. Parotid saliva protein profiles in caries-free and caries-active adults. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1997;83:244–51. doi: 10.1016/s1079-2104(97)90012-3. [DOI] [PubMed] [Google Scholar]
- 2.Banderas-Tarabay JA, Zacarías-D’Oleire IG, Garduño-Estrada R, Aceves-Luna E, González-Begné M. Electrophoretic analysis of whole saliva and prevalence of dental caries- A study in Mexican dental students. Arch Med Res. 2002;33:499–505. doi: 10.1016/s0188-4409(02)00395-8. [DOI] [PubMed] [Google Scholar]
- 3.Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, et al. Large scale identification of proteins in human salivary proteome by liquid chromatography-mass spectrometry and two dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–28. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 4.Oral health surveys: Basic methods. 3rd ed. Geneva, Switzerland: WHO; 1996. World Health Organizations. [Google Scholar]
- 5.FDI Working Group 10, Core: Saliva: its role in health and disease. Int Dent J. 1992;42:291–304. [PubMed] [Google Scholar]
- 6.Layne E. Spectrophotometric and turbidometric methods for measuring proteins. Meth Enzymol. 1957;3:447. [Google Scholar]
- 7.Schwartz SS, Zhu WX, Sreebny LM. Sodium do decyl sulphate polyacrylamide gel electrophoresis of human whole saliva. Archs Oral Biol. 1995;40:949–58. doi: 10.1016/0003-9969(95)00055-t. [DOI] [PubMed] [Google Scholar]
- 8.Mazengo MC, Tenovuo J, Hausen H. Dental caries in relation to diet, saliva and cariogenic micro-organisms in Tanzanians of selected age groups. Community Dent Oral Epidemiol. 1996;24:169–74. doi: 10.1111/j.1600-0528.1996.tb00836.x. [DOI] [PubMed] [Google Scholar]
- 9.Streckfus CF, Bigler LR. Saliva as a diagnostic fluid. Oral Dis. 2002;8:69–76. doi: 10.1034/j.1601-0825.2002.1o834.x. [DOI] [PubMed] [Google Scholar]
- 10.Johansson I, Saellström AK, Rajan BP, Parameswaran A. Salivary flow and dental caries in Indian children suffering from chronic malnutrition. Caries Res. 1992;26:38–43. doi: 10.1159/000261425. [DOI] [PubMed] [Google Scholar]
- 11.Leone CW, Oppenheim FG. Physical and Chemical aspects of saliva as indicators of risk for dental caries in humans. J Dent Educ. 2001;65:1054–62. [PubMed] [Google Scholar]
- 12.Swerdlove CK. Relation between the incidence of dental caries and the pH of normal resting saliva. J Dent Res. 1942;21:73–81. [Google Scholar]
- 13.de Farias DG, Bezerra AC. Salivary antibodies, amylase and protein from children with early childhood caries. Clin Oral Investig. 2003;7:154–7. doi: 10.1007/s00784-003-0222-7. [DOI] [PubMed] [Google Scholar]
- 14.Cohen RE, Aguirre A, Neiders ME, Levine MJ, Jones PC, Reddy MS, et al. Immunochemistry and immunogenicity of low- molecular weight human salivary mucin. Arch Oral Biol. 1991;36:347–56. doi: 10.1016/0003-9969(91)90004-E. [DOI] [PubMed] [Google Scholar]
- 15.Beeley JA. Clinical applications of electrophoresis of human salivary proteins. J Chromatogr. 1991;569:261–80. doi: 10.1016/0378-4347(91)80233-3. [DOI] [PubMed] [Google Scholar]
- 16.Tenovuo J. Salivary parameters of relevance for assessing caries activity in individuals and populations. Community Dent Oral Epidemiol. 1997;25:82–6. doi: 10.1111/j.1600-0528.1997.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 17.Kirstilä V, Häkkinen P, Jentsch H, Vilja P, Tenovuo J. Longitudinal analysis of the association of human salivary anti-microbial agents with caries increment and cariogenic micro-organisms: A two-year cohort study. J Dent Res. 1998;77:73–80. doi: 10.1177/00220345980770011101. [DOI] [PubMed] [Google Scholar]
- 18.Tenovuo J, Jentsch H, Soukka T, Karhuvaara L. Anti-microbial factors of saliva in relation to dental caries and salivary levels of mutans streptococci. J Biol Buccale. 1992;20:85–90. [PubMed] [Google Scholar]
- 19.Van Nieuw Amerongen A, Bolscher JG, Veerman EC. Salivary proteins: Protective and diagnostic value in cariology? Caries Res. 2004;38:247–53. doi: 10.1159/000077762. [DOI] [PubMed] [Google Scholar]


