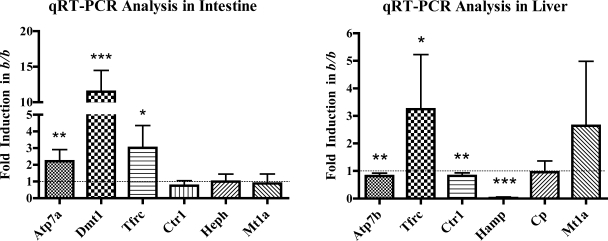
Fig. 3.
qRT-PCR analysis of intestinal and hepatic gene expression. qRT-PCR was performed with RNA samples extracted from isolated enterocytes (left) and liver (right) of +/b and b/b rats. Experimental repetitions utilizing different groups of +/b or b/b animals were as follows: +/b, n = 19 and b/b, n = 13 for intestine; +/b, n = 24 and b/b, n = 18 for liver. Y-axis shows fold change in b/bs compared with +/bs. The dashed line corresponding to 1.0-fold change (i.e., no change) on the y-axis is shown in both panels; bars below 1.0 indicate decreases, and bars above indicate increases in the b/b compared with the +/bs. *P < 0.05, **P < 0.01, ***P < 0.001; all indicating significant differences between genotypes. Means ± SD are shown. Atp7a, Menkes copper transporting ATPase; Atp7b, Wilson's copper transporting ATPase; Cp, ceruloplasmin; Ctr1, copper transporter 1; Dmt1, divalent metal transporter 1; Hamp, hepcidin; Heph, hephaestin; Mt1a, metallothionein 1A; Tfrc, transferrin receptor 1.