Background: Altered cell motility is a crucial aspect determining the invasiveness of tumors.
Results: Cells with concomitantly high levels of gangliosides and caveolin-1 have reduced activation of Src and reduced motility.
Conclusion: A caveolin-1·glycolipid complex can modulate the motility of tumor cells by regulating Src activity
Significance: This is a novel mechanism for the control of cell motility by caveolin-1 and gangliosides.
Keywords: Caveolin, Cell Motility, Ganglioside, Membrane Lipids, Sialyltransferase, Src
Abstract
The genetic (stable overexpression of sialyltransferase I, GM3 synthase) or pharmacological (selective pressure by N-(4-hydroxyphenyl)retinamide)) manipulation of A2780 human ovarian cancer cells allowed us to obtain clones characterized by higher GM3 synthase activity compared with wild-type cells. Clones with high GM3 synthase expression had elevated ganglioside levels, reduced in vitro cell motility, and enhanced expression of the membrane adaptor protein caveolin-1 with respect to wild-type cells. In high GM3 synthase-expressing clones, both depletion of gangliosides by treatment with the glucosylceramide synthase inhibitor d-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol and silencing of caveolin-1 by siRNA were able to strongly increase in vitro cell motility. The motility of wild-type, low GM3 synthase-expressing cells was reduced in the presence of a Src inhibitor, and treatment of these cells with exogenous gangliosides, able to reduce their in vitro motility, inactivated c-Src kinase. Conversely, ganglioside depletion by d-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol treatment or caveolin-1 silencing in high GM3 synthase-expressing cells led to c-Src kinase activation. In high GM3 synthase-expressing cells, caveolin-1 was associated with sphingolipids, integrin receptor subunits, p130CAS, and c-Src forming a Triton X-100-insoluble noncaveolar signaling complex. These data suggest a role for gangliosides in regulating tumor cell motility by affecting the function of a signaling complex organized by caveolin-1, responsible for Src inactivation downstream to integrin receptors, and imply that GM3 synthase is a key target for the regulation of cell motility in human ovarian carcinoma.
Introduction
Alterations in glycosphingolipid3 expression are common in tumors of different origins (2). Moreover, gangliosides, sialic acid-containing glycosphingolipids, are known to modulate several cellular functions relevant to tumor progression. Thus, altered ganglioside expression might play a relevant role in determining the aggressiveness and metastatic potential at least in certain tumors. Cellular ganglioside levels regulate cell proliferation, mainly by affecting the tyrosine kinase activity associated with growth factor receptors and receptor compartmentalization, thus regulating the responsiveness of these receptors to their ligands or their cross-talk with other signaling modules. However, gangliosides deeply affect tumor cell adhesion, motility, and migration. Ganglioside depletion by double knock-out of GM3 synthase and GM2 synthase in transformed fibroblasts reduced their migration on fibronectin and reduced their tumorigenic potential in syngeneic immunocompetent mice (3). However, transfection with GM1a/GD1b synthase, leading to the expression of complex gangliosides, inhibited neuroblastoma cell migration by affecting the Rho/Rac1 pathway (4). Invasiveness of melanoma cells was reduced by GM1/GD1b/GA1 synthase expression (5) or by GlcCer synthase antisense inhibition (6). GM3 is highly expressed in noninvasive compared with invasive bladder tumors and derived cell lines (7, 8); the overexpression of GM3 synthase reduced cell proliferation, motility, and invasion in mouse bladder carcinoma cells (9). The molecular mechanisms underlying the role of gangliosides in the regulation of tumor invasiveness are likely very heterogeneous. In colorectal (10) and bladder (8) cancer cells characterized by high GM3 levels and by the expression of tetraspanin CD9, a CD9·α3 integrin complex, were stabilized by GM3-mediated interactions. The Src C-terminal kinase CSK was recruited to this complex, with consequent inhibition of c-Src and reduced cell motility (11). Therefore, the influence of gangliosides on tumor cell motility seems to be mediated by the regulation of membrane-associated signaling complexes. Ganglioside interaction with hydrophobic membrane adaptor proteins seems to be crucial for this regulation. From this point of view, the interaction between gangliosides and the integral membrane protein caveolin-1 is potentially very interesting. Caveolin-1 is usually highly expressed in terminally differentiated cells, although it is markedly down-regulated in tumors of different origin, including ovarian, breast, and colon carcinoma (12). Cell lines with no caveolin-1 expression are usually characterized by increased proliferation (13), and in certain cell types, inhibition of caveolin-1 expression is sufficient to induce transformation and to accelerate tumorigenesis (14, 15), suggesting a role for caveolin-1 as a suppressor of tumor growth and metastasis in several types of cancer (16). In particular, caveolin-1 inhibits metastatic potential in melanoma through suppression of the integrin/Src/FAK signaling pathway (17).
In some cases, entire signaling modules are dynamically associated with caveolin-1, implying a major functional role for caveolin-1 as a molecular organizer for multiprotein signaling complexes. In particular, the amino acid sequence between 80 and 101 on the N-terminal side of the membrane insertion region of caveolin-1 is important for its interactions with many other proteins and has consequently been named “caveolin scaffolding domain” (18). Among others, caveolin-1 binds to nonreceptor kinases, including c-Src. Remarkably, for all of the enzymatic proteins that interact with caveolin-1 via a caveolin-binding motif, it has been observed that the binding motif is localized within the catalytic domain of the protein. In fact, in the case of tyrosine and serine/threonine kinases, the caveolin binding domain is localized within the catalytic kinase domain, and the caveolin scaffolding domain is able to inhibit the activity of several kinases, suggesting that the major regulatory function of caveolin-1 in multiprotein signaling complexes might rely on its ability to negatively affect multiple protein kinases associated with signaling modules (19). It is worthwhile to recall that many signaling modules are constitutively activated during cellular transformation. Among others, c-Src is usually overexpressed or activated in tumors. c-Src is a nonreceptor tyrosine kinase belonging to a family of proteins comprising nine members, c-Src, Fyn, Yes, Lck, Hck, Blk, Fgr, Lyn, and Yrk (20). In normal cells, Src family tyrosine kinases transduce signals acting on cell proliferation, adhesion, and motility (21). Particularly relevant is the role of Src in integrin-mediated cell adhesion (21, 22). Conversely, cell lines from metastatic tumors usually exhibit elevated Src kinase activity (23–27).
A relationship between c-Src and caveolin-1 has been documented in different cell types. Caveolin-1 interacts with c-Src within a glycosphingolipid-enriched membrane domain. The association between Src and caveolin is sensitive to the lipid composition of the membrane microenvironment (16, 28). Moreover, caveolin-1 reduces cell motility by inhibiting Src. For example, caveolin-1 inhibits metastatic potential in melanomas through the suppression of the integrin/Src/FAK signaling pathway (17). Caveolin-1 reduces osteosarcoma metastatic ability by inhibiting c-Src activity and Met signaling (29). In turn, Src can also contribute to caveolin-1-related signaling by regulating the phosphorylation levels of caveolin-1 (30, 31). In this study, we suggest that a ganglioside·caveolin-1 complex, controlling the activation of c-Src, might be involved in the regulation of human ovarian carcinoma cell motility.
EXPERIMENTAL PROCEDURES
Chemicals
Commercial chemicals were the purest available and, unless otherwise stated, were obtained by Sigma.
Lipids and Radioactive Lipids
Gangliosides and [1-3H]sphingosine were prepared as described previously (32).
Cell Culture
Cells (wild-type A2780 human ovarian carcinoma cells, the derived clonal line A2780/HPR4) were cultured in RPMI 1640 medium (Sigma) supplemented with 10% heat-inactivated FBS (Invitrogen), 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin in the presence of 5 μm HPR (33). Stably SAT-I-transfected A2780 cell lines were established and maintained as described previously (32).
Determination of in Vitro Cell Motility by Wound Healing Assay
Cells grown in 100-mm culture dishes as confluent monolayers were mechanically scratched using a 200-μl pipette tip, and wound healing was quantitatively assessed as described previously (32). For the determination of cell motility in caveolin-1-silenced cells, cells were first transfected with siRNA. After 48 h, cell monolayers were scratched, and transfection with siRNA was renewed as described below. Each experimental point was performed in duplicate, and data were expressed as the means ± S.D. of three independent experiments.
Determination of in Vitro Cell Motility by Phagokinetic Gold Sol Assay
Phagokinetic assays with gold colloid-coated plates were performed as described previously (34). Briefly, 24-mm coverslips were coated with 1% BSA (Sigma) and then immersed in the colloidal gold solution (35, 36). Two thousand cells were seeded on the gold colloid-coated coverslips and incubated at 37 °C. Images of the phagokinetic tracks were taken after different times by the use of a phase contrast microscope. The tracks of at least 50 cells were videocaptured, and the areas cleared from the gold colloid by cell phagocytosis, representing the migration response, were quantified by the use of Image J software. Data are expressed as the means ± S.D. of three independent experiments.
Western Blot
Cell homogenates, gradient fractions, and immunoprecipitation samples were analyzed by SDS-PAGE and immunoblotting using mouse polyclonal anti-caveolin-1, anti-p130CAS, anti-CD9, and anti-integrin α5 and β1 (BD Transduction Laboratories), rabbit monoclonal anti-c-Src, rabbit polyclonal anti-Src-pY416, and anti-Src-pY527 (Cell Signaling Technology), and mouse monoclonal anti-α-SMA (Sigma) followed by reaction with secondary horseradish peroxidase-conjugated antibodies and enhanced chemiluminescence detection (Pierce). β-Actin or β-tubulin was used as loading control (anti-β-actin rabbit polyclonal antibody was from Santa Cruz Biotechnology, Santa Cruz, CA; anti-β-tubulin monoclonal antibody was from Sigma). The data acquisition was performed using a GS-700 imaging densitometer, and acquired blots were elaborated using the Quantity One software (Bio-Rad). Each experimental point was performed in triplicate, and data were expressed as the means ± S.D. of three independent experiments.
RNA Extraction and RT-PCR
Total RNA extraction and semi-quantitative RT-PCR for CAV1, -2, and -3, has been performed as described previously (32). The housekeeping genes GAPDH and ACTB were used as reaction and loading control and were simultaneously amplified with the target genes. Data were acquired using a GelDoc 2000 instrument (Bio-Rad) and were elaborated using the Quantity One software (Bio-Rad). Data were normalized versus GAPDH expression and utilizing individual CAV1 and -2 of the A2780 specimen set as 1. Data are the means ± S.D. of three independent experiments.
Electron Microscopy
Cell cultures were processed and analyzed by transmission electron microscopy as described previously (32). Human skin fibroblasts have been used as the positive control for caveolae morphology.
siRNA Transfection
A2780/HPR or SAT-I-transfected cells were plated in 6-well plates or 100-mm dishes and, when grown at 50% confluence, were transfected with CAV1 siRNA (Qiagen, catalog no. SI00299635) and with scrambled siRNA duplexes (Qiagen, All Stars negative control siRNA catalog no. 1027280) as transfection control. The optimal condition for the transfection was 32 nm siRNA in OptiMEM with Lipofectamine 2000 (1%, v/v) (Invitrogen), following the protocol provided by the manufacturer. Fresh medium was added 24 h after transfection, and experiments were conducted for different times up to 72 h. In the case of the cell motility analysis by wound healing assay, cells were pretreated with siRNA for 48 h before the assay, and siRNA administration was repeated after 48 h.
Immunofluorescence Analysis of Caveolin-1
A2780 and A2780/HPR cells were grown on 24-mm sterilized coverslips. A2780/HPR cells were transfected with scrambled siRNA or siRNA targeting CAV1 mRNA as described above. After 72 h, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and stained with rabbit anti-caveolin-1 IgG diluted 1:5000 in phosphate-buffered saline supplemented with 1% BSA and a FITC-conjugated secondary antibody against rabbit IgG. The final samples were examined by fluorescence microscopy.
PDMP Treatment
To study the effects of ganglioside synthesis inhibition, the GlcCer synthase inhibitor d-PDMP has been used. As a negative control, cells were treated with the inactive stereoisomer l-PDMP under the same experimental conditions. d- and l-PDMP were kindly provided by Dr. Jin-ichi Inokuchi (Division of Glycopathology, Institute of Molecular Biomembranes and Glycobiology, Tohoku Pharmaceutical University, Aoba-ku, Sendai, Miyagi, Japan). The compounds were dissolved in distilled water at a concentration of 4 mm. The stock solution was stored at 4 °C and diluted with cell culture medium to a final concentration of 10 or 20 μm just before use. A2780/HPR cells were seeded and cultured in the presence of d- and l-PDMP for 48 h. The effects of PDMP on ganglioside synthesis were checked by analyzing the lipid composition of the treated cells as described below.
Treatment with Exogenous Gangliosides
Administration of exogenous gangliosides to A2780 cells has been performed as described previously (32).
Lipid Analysis
Cell sphingolipids were steady-state metabolically labeled by 2 h pulse/48 h chase with 3 × 10−8 m [1-3H]sphingosine as described previously (37). Lipids from total cell lysates or sucrose gradient fractions and immunoprecipitation samples obtained as described below were extracted with chloroform/methanol/water, 2:1:0.1, by volume (in the case of gradient fractions, water was omitted), subjected to a two-phase partitioning, and radioactive lipids were separated by monodimensional HPTLC (37) and quantitatively analyzed by digital autoradiography (37). Endogenous phospholipids, gangliosides, and cholesterol were analyzed as described previously (37).
Preparation of DRM Fractions by Sucrose Gradient Centrifugation
Cells were subjected to homogenization, lysis in the presence of Triton X-100, and ultracentrifugation on discontinuous sucrose gradients (32, 38). For sphingolipid analysis, cells were previously metabolically labeled with [1-3H]sphingosine as described above.
Immunoprecipitation Experiments
Aliquots of the detergent-resistant membrane fractions obtained from [1-3H]sphingosine-labeled cells were diluted 10-fold in immunoprecipitation buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm NaF, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, 1 mm PMSF, 75 milliunits/ml aprotinin, 1% Triton X-100). After preclearing for nonspecific binding, 10 μg/ml anti-caveolin-1 rabbit or 4 μg/ml anti-c-Src antibody or normal rabbit IgG (as negative control) was added to the supernatants, and the mixtures were put in rotation overnight at 4 °C. Immunocomplexes were recovered using protein A- and G-coupled magnetic beads (Dynabeads, Invitrogen) in rotation for 4 h at 4 °C. After delipidation by chloroform/methanol extraction, beads were suspended in Laemmli sample buffer and boiled 5 min at 100 °C, and the immunoprecipitated samples were separated from the beads by centrifugation.
Src Immunocomplex Kinase Assay
Cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm NaF, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 0.2% SDS, 0.5% sodium deoxycholate, 1 mm Na3VO4, 1 mm PMSF, 75 milliunits/ml aprotinin). Protein concentration was adjusted to 1 μg/μl with RIPA buffer. 500 μg of proteins from each sample were used for immunoprecipitation. Samples were pre-cleared for nonspecific binding by adding 10 μl of G protein-coupled magnetic beads (Dynabeads, Invitrogen). Precleared samples were incubated with 4 μg of anti-Src antibody (clone GD11, Millipore, Temecula, CA) or normal rabbit IgG, previously adsorbed on 10 μl of G protein-coupled beads following the manufacturer's instructions for 1 h at 4 °C. The anti-Src-immunoprecipitated samples, conjugated to the magnetic beads, were recovered and incubated with 10 μCi of [γ-32P]ATP (PerkinElmer Life Sciences) and 10 μm ATP in Kinase buffer (30 mm HEPES, pH 7.5, 10 mm MgCl2, 2 mm MnCl2, 0.2 mm Na3VO4, 2 mm NaF, 1 mm DTT, 1 mm PMSF, and 75 milliunits/ml aprotinin) at 37 °C for 1 h. The reaction was stopped by adding an equal volume of ice-cold 2× RIPA buffer, and beads were washed with ice-cold RIPA buffer and put at 100 °C for 5 min with 2× Laemmli buffer. The samples were separated by SDS-PAGE, and proteins were transferred to polyvinylidene difluoride membranes. Membranes were analyzed by autoradiography by exposing to Kodak BioMax MR Film (Sigma) at −80 °C. c-Src was detected by Western blot as described below.
Other Experimental Procedures
The protein content was determined according to Lowry et al. (39), using BSA as the reference standard. Radioactivity associated with cells, with medium, with gradient fractions, and with lipid extracts was determined by liquid scintillation counting.
Statistical Analysis
Experiments were performed in triplicate, unless otherwise stated. Data are expressed as means ± S.D. and were analyzed by one-way analysis of variance followed by the Student-Neuman-Keul's test. p values are indicated in the legend of each figure.
RESULTS
A2780/HPR Cells, Characterized by Up-regulation of GM3 Synthase, Are Less Motile than A2780 Cells
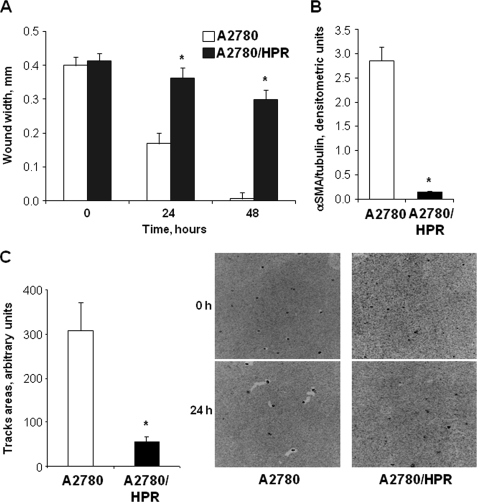
Previously, we showed that A2780/HPR cells, a clonal line obtained from human ovarian carcinoma A2780 cells by selective pressure in the presence of N-(4-hydroxyphenyl)-retinamide (HPR), are characterized by a marked up-regulation of GM3 synthase (sialyltransferase-I, SAT-I), a key enzyme in ganglioside biosynthesis. Consequently, A2780/HPR cells have higher levels of gangliosides with respect to the parental cell line (37). In addition, we showed that stable overexpression of GM3 synthase in A2780 cells led to reduced in vitro cell motility (32). To confirm a possible relationship between GM3 synthase expression and motility of ovarian cancer cells, we assessed the in vitro cell motility of A2780/HPR cells by wound healing assay and phagokinetic gold sol assay. Both assays revealed that the motility of A2780/HPR cells was strongly reduced when compared with wild-type A2780 cells (Fig. 1, panels A and C). In addition, we analyzed the expression of α-smooth muscle actin (α-SMA), a marker of the epithelium-to-mesenchyme transition and of myofibroblast activation, usually expressed at high levels in tumor cells with high ability to migrate and invade (40). We observed that α-SMA expression was strongly reduced in A2780/HPR (Fig. 1, panel B), as in SAT-I transfected A2780 cells (32), with respect to wild-type A2780 cells.
FIGURE 1.
In vitro motility of A2780 and A2780/HPR cells and α-SMA expression. Panel A, in vitro motility has been assessed in A2780 (white bars) and A2780/HPR cells (black bars) by wound healing assay. Confluent monolayers were wounded with a rubber policeman; phase contrast microscopy images of the wounds have been recorded, and the wound widths have been measured at 0, 24, and 48 h. Data are expressed in millimeters and are the means ± S.D. of three independent experiments. *, p < 0.01 versus time-matched A2780 cells. Panel B, protein levels of the epithelium-mesenchyme transition marker α-SMA have been evaluated on cell lysates from A2780 (white bar) and A2780/HPR cells (black bar) by Western blotting after SDS-PAGE separation. The amount of α-SMA present in each sample was determined by densitometry, normalized with respect to β-tubulin, used as loading control. Data are the means ± S.D. of three independent experiments. *, p < 0.01 versus A2780 cells. Panel C, in vitro motility of A2780 and A2780/HPR cells has been assessed by a phagokinetic gold sol assay. Briefly, cells were plated on colloidal gold-coated coverslips. Migrating cells ingest or displace colloidal gold particles, forming clear particle-free tracks. The areas of the tracks cleared by the cells have been recorded at time 0 and after 24 h. Average track areas (means ± S.D. of 50 measurements) normalized for the different cell size are reported in the left panel. *, p < 0.001 versus A2780 cells. Representative phase contrast microscopy images for each data set are shown in the right panel.
Caveolin-1 Is Markedly Up-regulated in A2780/HPR Cells
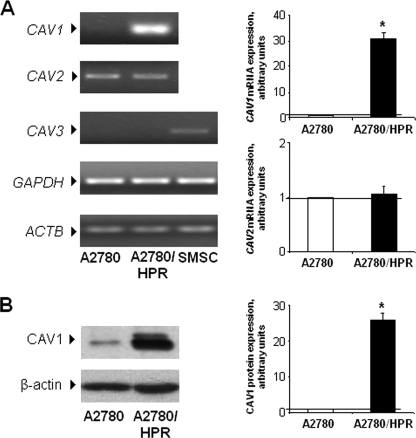
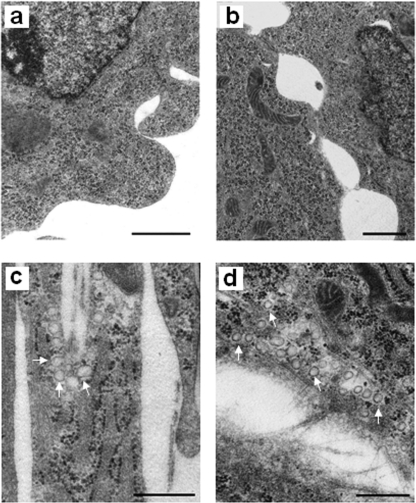
SAT-I-transfected A2780 cells are characterized by a strong up-regulation of caveolin-1 (32), a membrane adaptor protein that seems to act as a negative regulator of tumor progression and invasiveness (16). Thus, we checked caveolin expression in A2780/HPR cells, which are characterized by high GM3 synthase levels. The expression levels of caveolin family members (caveolin-1–3) were assessed in A2780 and A2780/HPR cells as mRNA by semi-quantitative RT-PCR (Fig. 2, panel A) and as protein by SDS-PAGE followed by Western blotting with specific antibodies (Fig. 2, panel B). Caveolin-1 was markedly up-regulated both at the mRNA and protein levels in A2780/HPR cells compared with A2780 cells, in which the protein is expressed at very low levels, in agreement with previously published data (41). Caveolin-2 and -3 were expressed at very similar levels in the two cell lines. Despite the major role of caveolin-1 as a structural component of caveolae (42), the up-regulation of caveolin-1 in A2780/HPR cells was not paralleled by the appearance of morphologically distinguishable caveolae (Fig. 3).
FIGURE 2.
Expression of caveolins in A2780 and A2780/HPR cells. Panel A, semi-quantitative PCR analysis of CAV1, -2, and -3 mRNA expression levels was performed in A2780 and A2780/HPR cells by simultaneous amplification of the housekeeping genes GAPDH and ACTB. Human skeletal muscle satellite cells (SMSC) have been used as positive control for CAV3 expression. Patterns (left) are representative of those obtained in three independent experiments. CAV3 signal was not detectable in A2780 and A2780/HPR cells. Right, in the graphs, data are normalized versus GAPDH expression and utilizing individual CAV1 and -2 of the A2780 specimen set as 1. Data are the means ± S.D. of three independent experiments. *, p < 0.01 versus A2780 cells. Panel B, caveolin-1 protein levels were assessed by Western blotting. Equal amounts of cellular proteins (corresponding to 30 μg) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were probed using specific anti-caveolin-1 and anti-β-actin monoclonal antibodies. Patterns (left) are representative of those obtained in three independent experiments. Right, in the graphs, the histogram represents mean densitometric quantification of caveolin-1. Data are normalized versus β-actin expression and utilizing caveolin-1 of the A2780 specimen set as 1. Data are the means ± S.D. of three independent experiments. *, p < 0.01 versus A2780 cells.
FIGURE 3.
Absence of caveolae in A2780/HPR cells. A2780/HPR cells (a and b) and human skin fibroblasts (c and d), used as positive control for caveolae morphology, were fixed, post-fixed, dehydrated, and embedded in Epoxy resin as described under “Experimental Procedures.” Ultramicrotome serrated sections were cut along transversal (a and c) and horizontal planes (b and d), stained with uranyl acetate and lead citrate, and examined by a Philips CM 10 transmission electron microscope. Scale bar, 500 nm. The white arrows indicate some caveolae in fibroblasts.
Caveolin-1 Silencing Increases the Motility of A2780/HPR and SAT-I-transfected A2780 Cells
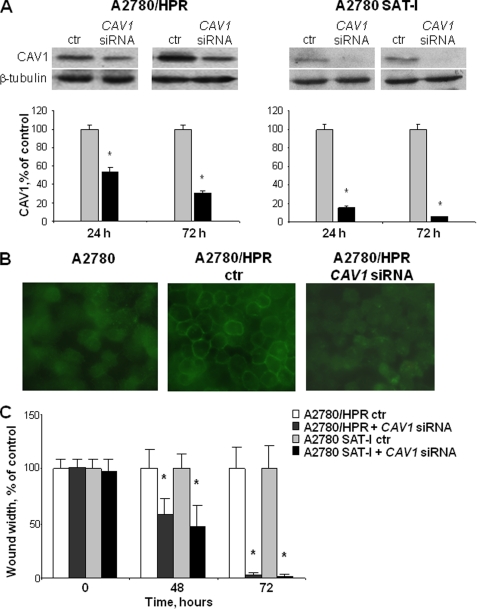
To verify whether caveolin-1 might act as a negative regulator of motility in cells with high GM3 synthase expression, caveolin-1 expression has been transiently knocked down using siRNA targeting CAV1 mRNA in A2780/HPR and SAT-I-transfected A2780 cells. The silencing efficacy was evaluated through Western blot analysis and immunofluorescence using a specific antibody for caveolin-1 (Fig. 4). siRNA treatment was able to significantly reduce caveolin-1 protein levels already after 24 h. After 72 h of treatment, caveolin-1 protein levels have been lowered by about 60 and 90% for A2780/HPR and SAT-I-transfected A2780 cells respectively, compared with the control cells transfected with scrambled siRNA duplex sequences (Fig. 4, panel A). The immunofluorescence analysis revealed that the immunoreactivity for caveolin-1 was almost completely lost from the plasma membrane in permeabilized CAV1 siRNA-treated A2780/HPR cells (Fig. 4, panel B). Similar results were obtained for SAT-I-transfected A2780 (data not shown). Caveolin-1 levels were unaffected in scrambled siRNA-treated cells. Caveolin-1 silencing had profound consequences on the motile ability of these cells. The in vitro motility of A2780/HPR and SAT-I-transfected A2780 cells, assessed by wound healing assay, was markedly higher in caveolin-1-silenced cells compared with control cells. A significant difference in cell motility was evident already after 48 h. Seventy two hours after the scratch, the caveolin-1 knocked down A2780/HPR and SAT-I-transfected A2780 cells completely healed the wound, suggesting a leading role for caveolin-1 in the regulation of the cell motility signal in this cell model (Fig. 4, panel C).
FIGURE 4.
Effects of caveolin-1 silencing on in vitro cell motility of A2780/HPR and SAT-I-transfected A2780 cells. A2780/HPR and SAT-I-transfected A2780 cells have been treated with siRNA targeting to CAV1 gene to reduce the expression of caveolin-1. Panel A, Western blot analysis and quantification of caveolin-1 expression in A2780/HPR and SAT-I-transfected A2780 cells treated with siRNA targeting to CAV1 mRNA (black bars) or with scrambled siRNA as transfection control (ctr) (gray bars). Analysis has been performed 24 and 72 h after siRNA administration. β-Tubulin was detected as a loading control. Patterns are representative of those obtained in three independent experiments (upper panel). The amount of caveolin-1 present in each sample was determined by densitometry, normalized respect to β-tubulin, and expressed as a percent of time-matched controls. Data are the means ± S.D. of three independent experiments. *, p < 0.01 versus time-matched controls (lower panel). Panel B, detection of caveolin-1 protein by immunofluorescence analysis. Untreated A2780 cells (left) and A2780/HPR cells treated with scrambled siRNA (middle) or with siRNA targeting to CAV1 (right) were permeabilized, exposed to a rabbit monoclonal anti-caveolin-1 antibody, and then incubated with a secondary anti-rabbit antibody conjugated to FITC as described under “Experimental Procedures.” Fluorescence images are representative of those obtained in three independent experiments. Panel C, effects of caveolin-1 silencing on in vitro cell motility of A2780/HPR and A2780 SAT-I cells. After 48 h treatment with scrambled siRNA or siRNA targeting to CAV1, wound healing assay was performed as described in the legend of Fig. 1, and siRNAs were maintained in the medium for the whole duration of the assay. Phase contrast images of the wounds were recorded at different times, and the wound widths were measured. Data are expressed in percentage respect to control cells treated with scrambled siRNA, and are the means ± S.D. of three independent experiments. *, p < 0.001 versus matched controls.
Blockade of Gangliosides Synthesis by GlcCer Synthase Pharmacological Inhibition Increases the Motility of A2780/HPR Cells
High expression levels of GM3 synthase, leading to high cellular gangliosides content, in both A2780/HPR and SAT-I-transfected A2780 cells were associated with a reduction of in vitro cell motility. Accordingly, treatment with exogenous gangliosides was able to reduce the motility of low GM3 synthase A2780 cells (32), suggesting a role of gangliosides in controlling the motility of these cells. To confirm this hypothesis, we assessed the effect of the pharmacological manipulation of ganglioside levels on the in vitro motility of A2780/HPR cells. Treatment of A2780/HPR cells with the specific GlcCer synthase inhibitor d-PDMP strongly reduced gangliosides content after 2 days and almost completely abolished GlcCer, LacCer, and ganglioside synthesis after 5 days (Fig. 5, panel A). l-PDMP (the inefficient isomer, used as a negative control) had no effect on glycosphingolipid levels in A2780/HPR cells. Treatment with l- and d-PDMP was not toxic; however, it slightly reduced cell proliferation in a dose-dependent manner (data not shown). However, treatment with d-PDMP, but not with l-PDMP, was able to significantly increase the motility of A2780/HPR cells, evaluated by the phagokinetic gold sol assay (Fig. 5, panel B). These results further support a role for gangliosides in the regulation of ovarian cancer cell motility.
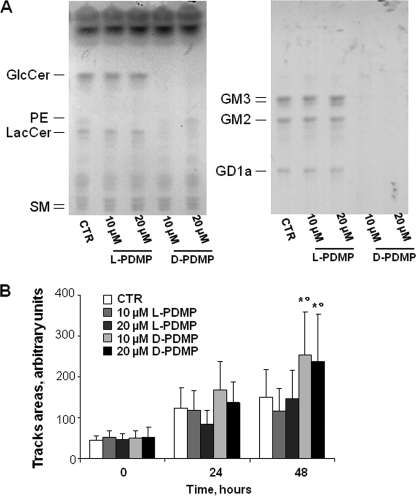
FIGURE 5.
Effects of PDMP treatment on the sphingolipid composition and on the in vitro cell motility of A2780/HPR cells. A2780/HPR cells were treated with the specific glucosylceramide synthase inhibitor d-PDMP to achieve sphingolipid depletion. The ineffective stereoisomer l-PDMP has been used as a negative control. Panel A, sphingolipid patterns of A2780/HPR cells untreated (CTR), treated with 10 μm and 20 μm l-PDMP or treated with 10 and 20 μm d-PDMP for 5 days. Cell lipids were extracted with chloroform/methanol/water, 2:1:0.1 by volume, subjected to a two-phase partitioning, and aqueous (left panel) and organic phases (right panel) lipids were analyzed by HPTLC, using as solvent system chloroform/methanol/water, 55:20:3 by volume (spray reagent, aniline/diphenylamine), and chloroform/methanol/0.2% aqueous CaCl2 50:42:11 (spray reagent, p-dimethylaminobenzaldehyde), respectively. The equivalent to 500 μg (for organic phases) and 1 mg (for aqueous phases) of cell proteins were loaded on each lane. PE, phosphatidylethanolamine. Panel B, effect of PDMP treatment on in vitro motility of A2780/HPR cells has been assessed by a phagokinetic gold sol assay as described in the legend of Fig. 1. A2780/HPR cells were treated with 10 or 20 μm of l- or d-PDMP for 48 h before the assay, and compounds were maintained in the medium for the whole duration of the assay. The areas of the tracks cleared by the cells have been recorded at time 0 and after 24 and 48 h. Average track areas (means ± S.D. of 50 measurements) normalized for the different cell size are reported in the bar graph. *, p < 0.002 versus time-matched control, °, p < 0.01 versus time-matched l-PDMP-treated cells.
Caveolin-1 Is Associated with Sphingolipids, Integrin Receptor Subunits, and c-Src in a Triton-insoluble Microenvironment
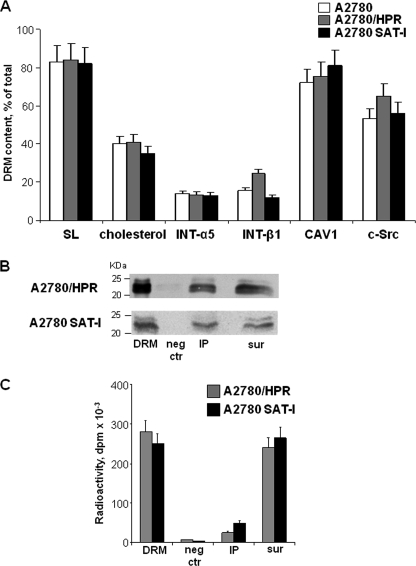
Altogether, these experimental data suggest that low motility in ovarian cancer cells is associated with high levels of both caveolin-1 and gangliosides. It has been suggested that caveolin-1 might act as a molecular organizer for membrane multiprotein signaling complexes, which are usually associated with Triton-insoluble, low buoyancy membrane fractions enriched in (glyco)sphingolipids and cholesterol. To ascertain the possible role of such a complex in controlling the motility of ovarian cancer cells, we prepared and analyzed DRM fractions from A2780, A2780/HPR, and SAT-I transfected A2780 cells using Triton X-100. DRM fractions from all the three cell types were similarly enriched in sphingolipids and cholesterol (about 80% of total cell sphingolipids and 40% of cholesterol were associated with DRM (Fig. 6, panel A)). DRM was highly enriched in caveolin-1 and c-Src (75% of total caveolin-1 and 66% of total c-Src were associated with DRM in A2780/HPR cells). No significant differences in the distribution of these antigens were observed in the different cell lines (Fig. 6, panel A). β1 and α5 integrin receptor subunits were also enriched in DRM, even if at a lesser extent (13.5% of total β1 integrin and 24.5% of total α5 integrin were recovered in the DRM in A2780/HPR cells.) No significant differences in the distribution of these antigens were observed in the different cell lines (Fig. 6, panel A), in agreement with previous reports indicating that these integrin receptors are relatively soluble in Triton X-100 (11, 43).
FIGURE 6.
Immunoprecipitation of caveolin-1 from a Triton X-100-resistant fraction from A2780, A2780/HPR, and SAT-1-transfected A2780 cells. Panel A, lipid and protein composition of the detergent-insoluble membrane fractions. After metabolic labeling of cell lipids with [1-3H]sphingosine, DRM fractions were prepared from A2780 (white bars), A2780/HPR (gray bars), and SAT-1-transfected A2780 cells (black bars) by sucrose gradient centrifugation after lysis in the presence of 1% Triton X-100 as described under “Experimental Procedures.” Lipids from the DRM fraction and from the other gradient fractions were extracted in chloroform/methanol and separated by HPTLC. Radioactive sphingolipids were detected and quantitatively analyzed by digital autoradiography, and cholesterol was detected colorimetrically and quantitatively analyzed by densitometry. Data are expressed as the percentage amount of total sphingolipids (SL) and cholesterol associated with the DRM fractions with respect to the total amount in the gradient. DRM and other gradient fractions have been also analyzed by SDS-PAGE followed by Western blotting detection using specific antibodies against β1 integrin, α5 integrin, caveolin-1, and c-Src. The relative quantities of each protein in the fraction were calculated by densitometry on the basis of the intensity of the blotting signal. Data are expressed as percentage of total signal associated with each antigen in the cell lysates. Data are the means ± S.D. of three independent experiments. Panel B, immunoprecipitation of caveolin-1 from DRM obtained from of A2780/HPR and SAT-I-transfected A2780 cells. Immunoprecipitation experiments using anti-caveolin-1 monoclonal antibody were performed starting from the same DRM amount under domain preserving conditions in which the interactions between lipid and protein within the plasma membrane have been maintained. Caveolin-1 was detected in each sample by Western blotting. 1st lane, DRM (1/30 of total volume). 2nd lane, negative control (neg ctr) (1/10 of the total volume). 3rd lane, anti-caveolin-1 immunoprecipitate (IP) (1/10 of the total volume). 4th lane, supernatants after immunoprecipitation (sur) (1/40 of total volume). Panel C, after metabolic labeling of cell sphingolipids with [1-3H]sphingosine, radioactivity-associated sphingolipids in immunoprecipitation samples obtained from of A2780/HPR and SAT-I-transfected A2780 cells (1st lane, whole DRM; 2nd lane, negative control; 3rd lane, anti-caveolin-1 immunoprecipitate; 4th lane, supernatants after immunoprecipitation) have been determined as described above. Data are expressed sphingolipid-associated radioactivity in each sample and are the means ± S.D. of three independent experiments.
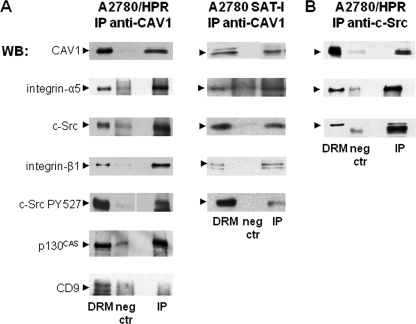
To study caveolin-1 microenvironment, we immunoprecipitated caveolin-1 from the DRM fractions obtained from A2780/HPR and SAT-I-transfected A2780 cells using anti-caveolin-1 monoclonal IgG under experimental conditions allowing the preservation of noncovalent interactions within DRM (44, 45). In both cases, the same amount of caveolin-1 was found associated with the immunoprecipitates (about 20% of total caveolin-1 present in the starting cell lysate) (Fig. 6, panel B). Conversely, caveolin-1 was quantitatively immunoprecipitated under experimental conditions allowing the complete antigen solubilization (data not shown). To allow the analysis of sphingolipid composition of anti-caveolin-1 immunoprecipitates, cell sphingolipids were steady-state metabolically labeled with [1-3H]sphingosine before DRM preparation and immunoprecipitation (37), and radioactive lipids associated with immunoprecipitation samples were extracted and quantitatively analyzed by two-dimensional TLC followed by digital autoradiography. A similar amount of sphingolipid-associated radioactivity was detected in anti-caveolin-1 immunoprecipitates (about 10% of total cell sphingolipids) from both A2780/HPR and SAT-I-transfected A2780 cells, with a neglectable amount of radioactivity associated with the immunoprecipitation negative controls (Fig. 6, panel C). No radioactive lipids were found in immunoprecipitates obtained after complete solubilization of caveolin-1 (data not shown). The sphingolipid pattern associated with the anti-caveolin-1-immunoprecipitated samples encompassed all the major sphingolipids present in these cells (GM3 and GM2 gangliosides, glucosylceramide, sphingomyelin, and ceramide) in the same proportions found in the starting DRM and in the total cell lysates (37). Thus, the environment of the caveolin-1 immunoseparated from DRM was characterized by a high lipid complexity. When immunoprecipitation experiments were performed under experimental conditions that allowed the preservation of noncovalent interactions, a complex pattern of proteins was co-immunoprecipitated with caveolin-1. The presence of α5 and β1 integrin receptor subunits, of c-Src (and its inactive form phosphorylated on tyrosine 527), and integrin signaling adaptor protein p130CAS in the anti-caveolin-1 immunoprecipitates from A2780/HPR and SAT-I-transfected A2780 cells was determined by immunoblotting using specific antibodies (Fig. 7, panel A). CD9, the only tetraspanin expressed in these cells, was not associated with the anti-caveolin-1 immunoprecipitates. To confirm the association of caveolin-1 with other DRM proteins, we performed immunoprecipitation experiments using an anti-c-Src monoclonal antibody. In the anti-c-Src immunoprecipitate, we were able to detect caveolin-1 and α5 integrin (Fig. 7, panel B), thus confirming the reciprocal association of caveolin-1 and c-Src in DRM.
FIGURE 7.
Proteins associated with the anti-caveolin-1 and anti-c-Src immunoprecipitates from a Triton-X-100-resistant fraction from A2780/HPR and SAT-1-transfected A2780 cells. Immunoprecipitation experiments using anti-caveolin-1 (panel A) or anti-c-Src (panel B) antibodies were performed starting from the same DRM amount under domain preserving conditions as described in the legend of Fig. 6. Caveolin-1, α5- and β1-integrin, c-Src, c-Src-pY527, CD9, and p130CAS were detected in each sample by Western blotting. 1st lane, DRM (1/30 of total volume). 2nd Lane, negative control (1/10 of the total sample volume) (neg ctr). 3rd lane, anti-caveolin-1 immunoprecipitate (panel A) or anti-c-Src immunoprecipitate (panel B) (1/10 of the total sample volume) (IP). Patterns are representative of those obtained in three different experiments.
c-Src Activation Is Negatively Regulated by Caveolin-1 and Gangliosides and Correlates with Cell Motility
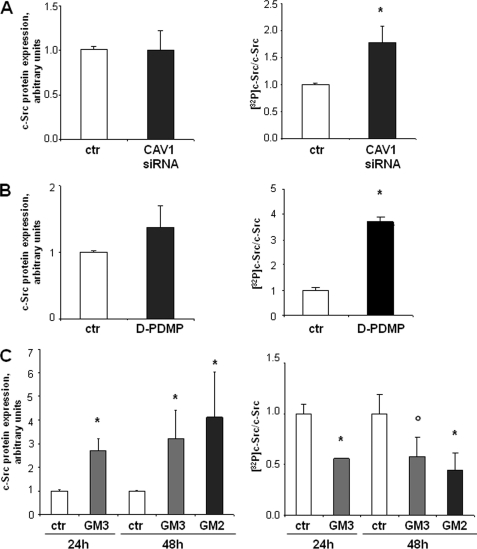
These results indicate that both caveolin-1 and gangliosides exert an inhibitory regulation on the motility of ovarian carcinoma cells. In addition, they suggest that gangliosides and caveolin-1 might be organized in a multimolecular complex together with c-Src and integrin receptors. Several papers indicated that GM3 is able to negatively affect cell motility stabilizing the formation of a integrin·tetraspanin membrane complex, leading to the inhibition of c-Src (10, 11, 46, 47). However, caveolin-1 suppression of metastatic potential in osteosarcoma (29) and melanoma (17) has been linked to the inhibition of c-Src activation. In addition, we have previously shown that the motility of low GM3 synthase-expressing A2780 cells was reduced in the presence of a Src inhibitor and that c-Src was less active in SAT-1-transfected cells (32). To confirm the role of c-Src in the control of cell motility in our model, we assessed Src kinase activity by immunocomplex kinase assay after different treatments were able to modify the gangliosides or caveolin-1 levels in these cells, resulting in variations in their motility. The results obtained are summarized in Fig. 8. Transient silencing of caveolin-1 in A2780/HPR cells (that positively affected in vitro cell motility) resulted in a higher c-Src kinase activity with respect to control cells treated with scrambled siRNA, not affecting the total cellular levels of c-Src (Fig. 8, panel A). In the same cells, abolition of ganglioside synthesis (associated with an increased cell motility) led as well to c-Src kinase activation with similar levels of c-Src protein (Fig. 8, panel B). However, the exogenous administration of monosialogangliosides GM3 and GM2 to A2780 cells (a treatment able to strongly inhibit their in vitro motility (32)) was able to significantly inhibit c-Src autophosphorylation, even if in this case we observed an increased amount of c-Src protein in the ganglioside-treated samples, which might represent a compensatory mechanism (Fig. 8, panel C). These data strongly suggest that the activity of c-Src, which is inversely correlated with cell motility, is regulated by the membrane ganglioside composition and/or the presence of caveolin-1.
FIGURE 8.
Effects of the modulation of caveolin-1 and cellular ganglioside levels on c-Src activity. c-Src activity has been measured as autophosphorylation using an immunocomplex kinase assay as described under “Experimental Procedures.” Briefly, c-Src has been immunoprecipitated with a specific monoclonal antibody from cell lysates obtained under the different experimental conditions. Immunoprecipitates have been incubated with 10 μm [γ-32P]ATP. After incubation, proteins associated with each sample have been separated by SDS-PAGE. The total amount of c-Src associated with each sample has been determined by Western blotting and densitometry (left panels). The amount of radioactivity associated with the band corresponding to c-Src protein has been determined by autoradiography and has been normalized for the total c-Src content associated with each sample (right panels). Data are the means ± S.D. of four independent experiments. Panel A, effect of caveolin-1 silencing in A2780/HPR cells. A2780/HPR cells were treated with siRNA targeting to CAV1 mRNA (gray bars) or with scrambled siRNA as control (white bars). Analysis has been performed 48 h after siRNA administration. *, p < 0.03 versus control. Panel B, effect of ganglioside depletion by treatment with d-PDMP in A2780/HPR cells. A2780/HPR cells were treated with 20 μm d-PDMP (gray bars) or with vehicle (white bars) for 48 h. *, p < 0.02 versus control. Panel C, effect of exogenous gangliosides administration in A2780 cells. Cells were treated with 50 μm GM2 for 48 h or with 50 μm GM3 for 24 and 48 h in serum-free medium as described under “Experimental Procedures.” *, p < 0.01 and °, p < 0.03 versus time-matched controls.
DISCUSSION
The evidence provided in this paper, together with our previously published results (32), indicate that, in human ovarian cancer cells, an increased cellular gangliosides content, caused by the enhanced expression of GM3 synthase, is paralleled by a marked up-regulation of the membrane adaptor protein caveolin-1. The result of concomitantly high levels of gangliosides and caveolin-1 is a marked reduction of in vitro cell motility. Up-regulation of caveolin-1 and reduced cell motility were observed in two cellular models in which higher GM3 synthase activity with respect to wild-type cells has been obtained by two entirely different procedures as follows: the genetic manipulation of GM3 synthase levels by its stable overexpression in the case of SAT-I-transfected A2780 cells or the selective pressure in the presence of a drug, N-(4-hydroxyphenyl)retinamide), in the case of A2780/HPR cells. The existence of a causal connection between high levels of gangliosides and caveolin-1 and the observed reduction of cell motility are supported by the depletion experiments, as shown in Figs. 4 and 5. Both caveolin-1 silencing and block of gangliosides biosynthesis by pharmacological inhibition of glucosylceramide synthase led to a strong enhancement of cell motility. These results indicate that high levels of caveolin-1 and high levels of gangliosides are necessary but not sufficient, if independent, to down-regulate cell motility. Thus, the regulation of cell motility requires a certain degree of cooperation between gangliosides and caveolin-1. Caveolin-1, an integral membrane protein originally discovered as a main structural component of caveolae, soon gained a role as a molecular organizer for multiprotein signaling complexes, because of its ability to interact with several proteins involved in signal transduction and to concentrate whole signaling modules in specialized plasma membrane areas, allowing their functional regulation. Caveolin-1 is insoluble in cold nonionic detergents (48) and can be enriched in low density Triton X-100-insoluble membrane fractions. These fractions, highly enriched also in cholesterol and sphingolipids, putatively correspond to lipid rafts. Thus, caveolin-1 at the plasma membrane is concentrated in a lipid-rich membrane environment, and lipids affect several of the functionally relevant properties of caveolin-1. Caveolin-1 and sphingolipids not only co-localize in the same detergent-insoluble membrane fraction, but several pieces of evidence indicate that they can be close enough in specialized membrane subdomains to allow a direct interaction between the transmembrane domain of caveolin and the hydrophobic moiety of the lipid (49, 50). In SAT-I-transfected A2780 ovarian carcinoma cells, photoreactive GM3 is able to label caveolin-1 in a detergent-resistant membrane preparation (32). In a few cases, it has also been reported that detergent-resistant association of caveolin-1 and sphingolipids is strong enough to allow co-immunoprecipitation. Caveolin-1 can be immunoprecipitated by a monoclonal antibody to ganglioside GD3 in CHO cells transiently transfected by GD3 synthase cDNA (51).
Here, we report that caveolin-1 and gangliosides are enriched in a Triton-X-100-insoluble fraction and that a significant portion of the sphingolipids associated with the DRM fraction can be recovered upon immunoprecipitation with anti-caveolin-1 antibody (Fig. 6). Therefore, we hypothesize that the formation of a ganglioside·caveolin-1 complex, occurring in cells with concomitantly high levels of both components, might be involved in the negative regulation of ovarian carcinoma cell motility. We tried to elucidate the downstream signaling pathway possibly affected by the ganglioside·caveolin-1 complex. Our results indicated that integrin receptor subunits and the nonreceptor tyrosine kinase c-Src are enriched together with caveolin-1 and sphingolipids within detergent-resistant membrane fractions. Moreover, integrin receptor subunits and c-Src co-immunoprecipitate with caveolin-1. Caveolin-1 (16, 28) and Src kinases (52, 53) are typically associated with sphingolipid-enriched membrane domains or other subtypes of lipid rafts also in other cell types. In addition, it has been suggested that caveolin-1 might act as a membrane adaptor coupling integrin receptors to Src kinases (54) and that caveolin-1-mediated inactivation of the integrin/Src/FAK pathway might be responsible for the inhibition of metastatic potential in melanoma (17). Our results strongly support the hypothesis that the inactivation of c-Src by a ganglioside·caveolin-1 complex might result in the down-regulation of ovarian carcinoma cell motility. We have previously shown that a Src inhibitor was able to inhibit the motility of A2780 cells (expressing low GM3 synthase and low caveolin-1) and that c-Src was less active in SAT-I-transfected cells (32), characterized by high caveolin-1 levels. Now we report that treatments able to reduce the content of caveolin-1 (CAV1 siRNA) or of gangliosides (glucosylceramide synthase inhibition) in A2780/HPR and in SAT-I-transfected A2780 cells led to enhanced c-Src kinase activation and increased motility. However, ganglioside administration to A2780 cells, which able to reduce their motility, resulted in c-Src kinase inactivation. Altogether, these data suggest a novel role for gangliosides in the regulation of cell motility in human ovarian carcinoma by affecting the organization of a signaling complex organized by caveolin-1, responsible for Src inactivation.
Our results call attention to two aspects that probably deserve future consideration. First, despite the much higher expression of caveolin-1, the morphological analysis revealed the absence of caveolae both in A2780/HPR and SAT-I-transfected A2780 cells as in the wild-type A2780 cells. This finding might seem in contrast to the usual association between caveolin-1 and caveolae, but it is indeed in agreement with the diverse and multiple caveolae-independent roles of caveolin-1 that have been described in the last few years (16, 28, 55). The second aspect is related to the observation that cellular ganglioside or GM3 synthase levels can regulate the expression of caveolin-1. Yamagata and co-workers (56) showed that GD1a ganglioside regulated caveolin-1 expression in FBJ mouse osteosarcoma cells. Also in that case, the concomitant increase in a certain ganglioside and in caveolin-1 seems to be related to the possible role of caveolin-1 as a tumor suppressor. FBJ mouse osteosarcoma cells and A2780 human ovarian carcinoma cells exist in different phenotypic variants, characterized by strikingly different in vitro cell motility. For both cell types, the low motility variants are characterized by high ganglioside and high caveolin-1 expression, although the high motility variants contain low ganglioside and low caveolin-1 levels. Treatment of the highly motile FBJ-LL osteosarcoma cell line with exogenous GD1a ganglioside or transfection with GM2/GD2 synthase cDNA resulted in the up-regulation of caveolin-1 expression with reduced metastatic potential and suppressed cell adhesion to vitronectin (57). Similarly, GM3 synthase-transfected A2780 ovarian carcinoma cells were characterized by an increased expression of caveolin-1 and reduced in vitro cell motility. However, the mechanism underlying the regulation of caveolin-1 expression by the cellular ganglioside levels is totally unknown.
Ganglioside and glycosphingolipid nomenclature is in accordance with the IUPAC-IUBMB recommendations (1).
- HPR
- N-(4-hydroxyphenyl)retinamide
- PDMP
- threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol
- DRM
- detergent-resistant membrane fraction
- EGFR
- epidermal growth factor receptor
- HPTLC
- high performance thin layer chromatography
- α-SMA
- α-smooth muscle actin.
REFERENCES
- 1. IUPAC-IUBMB, Joint Commission on Biochemical Nomenclature (1998) Carbohydr. Res. 312, 167–175 [Google Scholar]
- 2. Hakomori S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10231–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y., Yan S., Wondimu A., Bob D., Weiss M., Sliwinski K., Villar J., Notario V., Sutherland M., Colberg-Poley A. M., Ladisch S. (2010) Oncogene 29, 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dong L., Liu Y., Colberg-Poley A. M., Kaucic K., Ladisch S. (2011) Glycoconj. J. 28, 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong Y., Ikeda K., Hamamura K., Zhang Q., Kondo Y., Matsumoto Y., Ohmi Y., Yamauchi Y., Furukawa K., Taguchi R., Furukawa K. (2010) Cancer Sci. 101, 2039–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deng W., Li R., Guerrera M., Liu Y., Ladisch S. (2002) Glycobiology 12, 145–152 [DOI] [PubMed] [Google Scholar]
- 7. Kawamura S., Ohyama C., Watanabe R., Satoh M., Saito S., Hoshi S., Gasa S., Orikasa S. (2001) Int. J. Cancer 94, 343–347 [DOI] [PubMed] [Google Scholar]
- 8. Satoh M., Ito A., Nojiri H., Handa K., Numahata K., Ohyama C., Saito S., Hoshi S., Hakomori S. I. (2001) Int. J. Oncol. 19, 723–731 [PubMed] [Google Scholar]
- 9. Watanabe R., Ohyama C., Aoki H., Takahashi T., Satoh M., Saito S., Hoshi S., Ishii A., Saito M., Arai Y. (2002) Cancer Res. 62, 3850–3854 [PubMed] [Google Scholar]
- 10. Ono M., Handa K., Sonnino S., Withers D. A., Nagai H., Hakomori S. (2001) Biochemistry 40, 6414–6421 [DOI] [PubMed] [Google Scholar]
- 11. Mitsuzuka K., Handa K., Satoh M., Arai Y., Hakomori S. (2005) J. Biol. Chem. 280, 35545–35553 [DOI] [PubMed] [Google Scholar]
- 12. Wiechen K., Diatchenko L., Agoulnik A., Scharff K. M., Schober H., Arlt K., Zhumabayeva B., Siebert P. D., Dietel M., Schäfer R., Sers C. (2001) Am. J. Pathol. 159, 1635–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee S. W., Reimer C. L., Oh P., Campbell D. B., Schnitzer J. E. (1998) Oncogene 16, 1391–1397 [DOI] [PubMed] [Google Scholar]
- 14. Engelman J. A., Zhang X. L., Razani B., Pestell R. G., Lisanti M. P. (1999) J. Biol. Chem. 274, 32333–32341 [DOI] [PubMed] [Google Scholar]
- 15. Williams T. M., Medina F., Badano I., Hazan R. B., Hutchinson J., Muller W. J., Chopra N. G., Scherer P. E., Pestell R. G., Lisanti M. P. (2004) J. Biol. Chem. 279, 51630–51646 [DOI] [PubMed] [Google Scholar]
- 16. Prinetti A., Prioni S., Loberto N., Aureli M., Chigorno V., Sonnino S. (2008) Biochim. Biophys. Acta 1780, 585–596 [DOI] [PubMed] [Google Scholar]
- 17. Trimmer C., Whitaker-Menezes D., Bonuccelli G., Milliman J. N., Daumer K. M., Aplin A. E., Pestell R. G., Sotgia F., Lisanti M. P., Capozza F. (2010) Cancer Res. 70, 7489–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li S., Couet J., Lisanti M. P. (1996) J. Biol. Chem. 271, 29182–29190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smart E. J., Graf G. A., McNiven M. A., Sessa W. C., Engelman J. A., Scherer P. E., Okamoto T., Lisanti M. P. (1999) Mol. Cell. Biol. 19, 7289–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomas S. M., Brugge J. S. (1997) Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 21. Playford M. P., Schaller M. D. (2004) Oncogene 23, 7928–7946 [DOI] [PubMed] [Google Scholar]
- 22. Cary L. A., Klinghoffer R. A., Sachsenmaier C., Cooper J. A. (2002) Mol. Cell. Biol. 22, 2427–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Irby R. B., Yeatman T. J. (2000) Oncogene 19, 5636–5642 [DOI] [PubMed] [Google Scholar]
- 24. Hakak Y., Martin G. S. (1999) Curr. Biol. 9, 1039–1042 [DOI] [PubMed] [Google Scholar]
- 25. Verbeek B. S., Vroom T. M., Adriaansen-Slot S. S., Ottenhoff-Kalff A. E., Geertzema J. G., Hennipman A., Rijksen G. (1996) J. Pathol. 180, 383–388 [DOI] [PubMed] [Google Scholar]
- 26. Egan C., Pang A., Durda D., Cheng H. C., Wang J. H., Fujita D. J. (1999) Oncogene 18, 1227–1237 [DOI] [PubMed] [Google Scholar]
- 27. Harris K. F., Shoji I., Cooper E. M., Kumar S., Oda H., Howley P. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sonnino S., Prinetti A. (2009) FEBS Lett. 583, 597–606 [DOI] [PubMed] [Google Scholar]
- 29. Cantiani L., Manara M. C., Zucchini C., De Sanctis P., Zuntini M., Valvassori L., Serra M., Olivero M., Di Renzo M. F., Colombo M. P., Picci P., Scotlandi K. (2007) Cancer Res. 67, 7675–7685 [DOI] [PubMed] [Google Scholar]
- 30. Park J. H., Ryu J. M., Han H. J. (2011) J. Cell Physiol. 226, 267–275 [DOI] [PubMed] [Google Scholar]
- 31. Banquet S., Delannoy E., Agouni A., Dessy C., Lacomme S., Hubert F., Richard V., Muller B., Leblais V. (2011) Cell. Signal. 23, 1136–1143 [DOI] [PubMed] [Google Scholar]
- 32. Prinetti A., Aureli M., Illuzzi G., Prioni S., Nocco V., Scandroglio F., Gagliano N., Tredici G., Rodriguez-Menendez V., Chigorno V., Sonnino S. (2010) Glycobiology 20, 62–77 [DOI] [PubMed] [Google Scholar]
- 33. Illuzzi G., Bernacchioni C., Aureli M., Prioni S., Frera G., Donati C., Valsecchi M., Chigorno V., Bruni P., Sonnino S., Prinetti A. (2010) J. Biol. Chem. 285, 18594–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Albrecht-Buehler G. (1977) Cell 11, 395–404 [DOI] [PubMed] [Google Scholar]
- 35. Zetter B. R. (1987) Methods Enzymol. 147, 135–144 [DOI] [PubMed] [Google Scholar]
- 36. Scott W. N., McCool K., Nelson J. (2000) Anal. Biochem. 287, 343–344 [DOI] [PubMed] [Google Scholar]
- 37. Prinetti A., Basso L., Appierto V., Villani M. G., Valsecchi M., Loberto N., Prioni S., Chigorno V., Cavadini E., Formelli F., Sonnino S. (2003) J. Biol. Chem. 278, 5574–5583 [DOI] [PubMed] [Google Scholar]
- 38. Prinetti A., Chigorno V., Tettamanti G., Sonnino S. (2000) J. Biol. Chem. 275, 11658–11665 [DOI] [PubMed] [Google Scholar]
- 39. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 40. Yang J., Liu Y. (2001) Am. J. Pathol. 159, 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hinrichs J. W., Klappe K., Hummel I., Kok J. W. (2004) J. Biol. Chem. 279, 5734–5738 [DOI] [PubMed] [Google Scholar]
- 42. Anderson R. G. (1998) Annu. Rev. Biochem. 67, 199–225 [DOI] [PubMed] [Google Scholar]
- 43. Kazui A., Ono M., Handa K., Hakomori S. I. (2000) Biochem. Biophys. Res. Commun. 273, 159–163 [DOI] [PubMed] [Google Scholar]
- 44. Loberto N., Prioni S., Bettiga A., Chigorno V., Prinetti A., Sonnino S. (2005) J. Neurochem. 95, 771–783 [DOI] [PubMed] [Google Scholar]
- 45. Rivaroli A., Prioni S., Loberto N., Bettiga A., Chigorno V., Prinetti A., Sonnino S. (2007) J. Neurochem. 103, 1954–1967 [DOI] [PubMed] [Google Scholar]
- 46. Kawakami Y., Kawakami K., Steelant W. F., Ono M., Baek R. C., Handa K., Withers D. A., Hakomori S. (2002) J. Biol. Chem. 277, 34349–34358 [DOI] [PubMed] [Google Scholar]
- 47. Miura Y., Kainuma M., Jiang H., Velasco H., Vogt P. K., Hakomori S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16204–16209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kurzchalia T. V., Dupree P., Parton R. G., Kellner R., Virta H., Lehnert M., Simons K. (1992) J. Cell Biol. 118, 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chigorno V., Palestini P., Sciannamblo M., Dolo V., Pavan A., Tettamanti G., Sonnino S. (2000) Eur. J. Biochem. 267, 4187–4197 [DOI] [PubMed] [Google Scholar]
- 50. Fra A. M., Masserini M., Palestini P., Sonnino S., Simons K. (1995) FEBS Lett. 375, 11–14 [DOI] [PubMed] [Google Scholar]
- 51. Kasahara K., Watanabe Y., Yamamoto T., Sanai Y. (1997) J. Biol. Chem. 272, 29947–29953 [DOI] [PubMed] [Google Scholar]
- 52. Bénistant C., Chapuis H., Mottet N., Noletti J., Crapez E., Bali J. P., Roche S. (2000) Biochem. Biophys. Res. Commun. 273, 425–430 [DOI] [PubMed] [Google Scholar]
- 53. Bénistant C., Bourgaux J. F., Chapuis H., Mottet N., Roche S., Bali J. P. (2001) Cancer Res. 61, 1415–1420 [PubMed] [Google Scholar]
- 54. Wary K. K., Mariotti A., Zurzolo C., Giancotti F. G. (1998) Cell 94, 625–634 [DOI] [PubMed] [Google Scholar]
- 55. Head B. P., Insel P. A. (2007) Trends Cell Biol. 17, 51–57 [DOI] [PubMed] [Google Scholar]
- 56. Wang L., Takaku S., Wang P., Hu D., Hyuga S., Sato T., Yamagata S., Yamagata T. (2006) Glycoconj. J. 23, 303–315 [DOI] [PubMed] [Google Scholar]
- 57. Hyuga S., Yamagata S., Takatsu Y., Hyuga M., Nakanishi H., Furukawa K., Yamagata T. (1999) Int. J. Cancer 83, 685–691 [DOI] [PubMed] [Google Scholar]