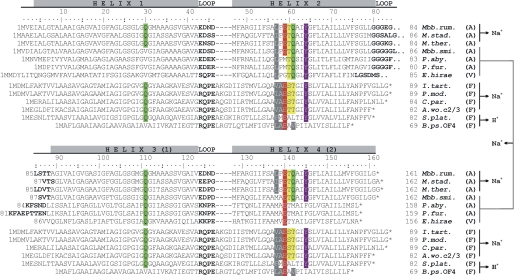
FIGURE 7.
Alignment of the rotor ring subunits (c/K) from A-, V-, and F-type ATPases/synthases. The chains consist either of 1 (F-type) or 2 (A- and V-type) α-helical hairpins (α-helix loop-α-helix motif). The residues involved in Na+ coordination (47–49) or H+ coordination (62, 63) are given in colors. The c subunit of M. ruminantium M1 harbors two hairpins, each displaying one complete Na+-binding signature. The signature is equivalent to the E. hirae Na+ binding signature (1 Na+-binding site per c/K subunit consisting of 4 α-helices), which involves in this case three α-helices. Residue numbering (according to the M. ruminantium gene sequence) and α-helices are indicated. Loop regions are in boldface. Residue numbers of individual sequences are given at the beginning and end of the sequences, and the asterisk indicates the sequence end. The ion-binding type (Na+ or H+), the type of ATPase/synthase (A, V, and F), and the involvement of which hairpins in ion binding for the individual sequences is indicated on the right side after the species name. Sources of amino acid sequences were as follows: M. ruminantium M1 (Mbb. rum.; accession number YP_003423440), Methanosphaera stadtmanae (M. stad.; YP_448163), M. thermautotrophicus (M. ther.; NP_276094), Methanobrevibacter smithii (Mbb. smi.; YP_001273012), Pyrococcus abyssi (P. aby.; NP_127441), Pyrococcus furiosus (P. fur.; NP_577907), E. hirae (E. hirae; BAA04273), I. tartaricus (I. tart.; AAM94908.1), P. modestum (P. mod.; CAA41369.1), Clostridium paradoxum (C. par.; ABB13420.1), A. woodii (A. wo. c2/3; AAC45088.2/AAF01475.1), Spirulina platensis (S. plat.; EF520738), and Bacillus pseudofirmus OF4 (B. ps. OF4; YP_003426325.1).