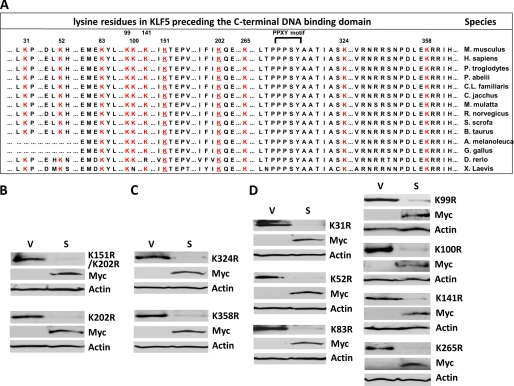
FIGURE 7.
Mutation at each lysine residue preceding the DNA binding domain of KLF5 does not prevent SMURF2 from degrading KLF5. A, alignment of all the lysine residues preceding the DNA binding domain of KLF5 from various species. Numbers indicate positions of these lysine residues, which are shown in red. Conserved residues adjacent to these lysines are also shown. The two SUMOylation site lysine residues are underlined (18). The PPXY (PPPSY) motif is bracketed (5). B, the two SUMOylation site lysines are not targets of SMURF2. HEK293T cells were transfected with the indicated KLF5 SUMOylation mutants and either Myc-SMURF2 (S) or vector alone (V), and whole cell lysates were subjected to Western blotting with mouse HA, Myc, and β-actin antibodies. K, lysine; r = arginine, Myc, Myc-SMURF2. C, individual mutations at Lys-324 and Lys-358, the two lysine lysines adjacent to the PY motif of KLF5, do not block the degradation KLF5 by SMURF2. HEK293T cells were transfected with K324R or K358R and either Myc-SMURF2 or vector alone, and whole cell lysates were subjected to Western blotting. D, mutation at each of the other lysine residues at the N terminus of KLF5 does not block the degradation of KLF5 by SMURF2. HEK293T cells were transfected with each indicated mutant and either Myc-SMURF2 or vector alone and whole cell lysates subjected to Western blotting.