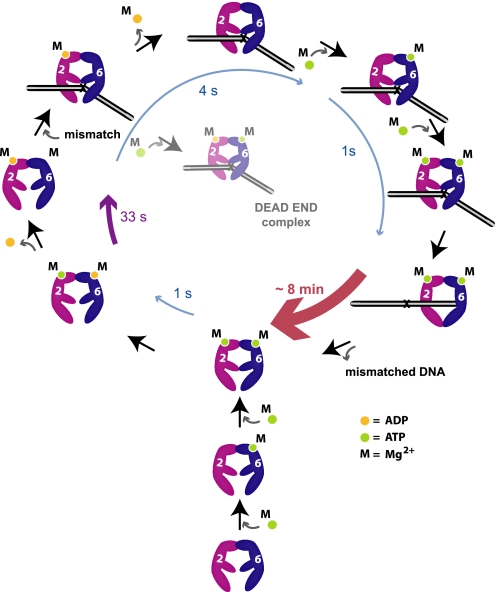
FIGURE 5.
An illustration of the kinetic events associated with hMSH2 and hMSH6 subunits during ADP/ATP processing for MMR. Bottom and lower left of cycle, hMSH6 rapidly binds ATP (green dot) followed by rapid binding of ATP to hMSH2. hMSH6 rapidly hydrolyzes ATP and releases the resultant ADP (yellow dot). hMSH2 hydrolyzes ATP more slowly yet retains ADP. This process results in the major steady-state species hMSH2(ADP)-hMSH6(apo). The likely disposition of magnesium (M) for each subunit is shown. Remaining cycle, upon mismatch interaction, hMSH2 rapidly releases ADP (4 s). Under conditions where hMSH6 is bound to ATP at the mismatch prior to ADP release, a dead-end complex is formed that cannot undergo nucleotide exchange. hMSH6 rapidly binds ATP, which enhances ATP binding by hMSH2 (1 s). Binding of ATP by both hMSH2 and hMSH6 results in the formation of a stable sliding clamp (∼8 min) that dissociates from the mismatch to execute the downstream steps of MMR. Dissociation from the DNA containing the mismatch and ATP hydrolysis completes the hMSH2-hMSH6 ATPase cycle.