FIGURE 2.
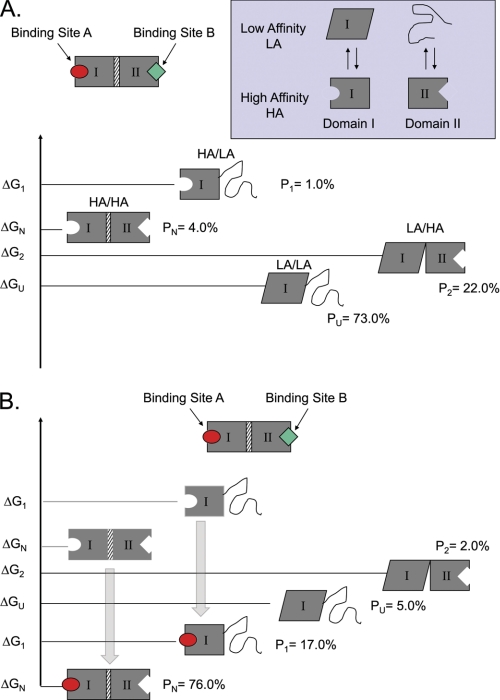
Tuning the protein ensemble can modulate allosteric control. A simple two-domain model can be used to demonstrate how changing the probabilities of states can result in differential allosteric effects. A, a two-domain protein is shown, with the domains connected through an interface. Each domain has an equilibrium between a HA and LA state, which is governed by the intrinsic stability terms (ΔGI and ΔGII). The LA state can be ID (as with the NTDs of the SHRs) or just an alternative conformation (as with the DBDs of SHRs). Coupling between domains is facilitated by an interaction energy (Δgint), which determines the energetic cost of having the HA states populated in each domain simultaneously. B, Le Chatelier's principle dictates that the addition of ligand will stabilize the HA state of a particular domain, causing a ligand-induced redistribution of the ensemble. In this example, the addition of ligand A stabilizes the HA state of domain I, and the resultant redistribution also stabilizes the HA state for domain II. The observed effect is an apparent increase in the affinity of the protein for ligand B (i.e. ligand A is a positive allosteric effector of ligand B). However, the impact that such a redistribution will have on the affinity of the protein for a second ligand will depend on where the equilibrium is poised prior to stimulation with ligand A. The probabilities (P) of each form under specific sets of conditions are shown. When this model was exhaustively explored, it was found that ID could be used to tune the allosteric response of a protein, with some parameter combinations producing positive coupling and others producing negative coupling (91).