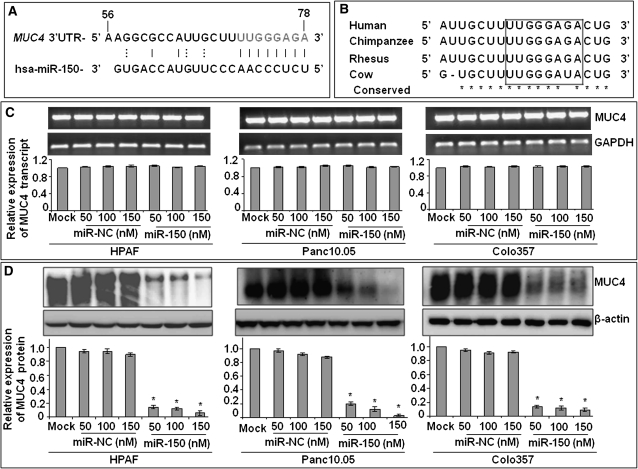
Fig. 1.
miR-150 negatively regulates the expression of MUC4. (A) Identification of a putative miR-150-binding site in the MUC4 3′ UTR at position. Eight bases (71 through 78) of the MUC4 3′ UTR are perfect matches (seed sequence) for the miR-150. (B) Comparison of the MUC4-binding element among mammals demonstrates a high degree of conservation. (C and D) Posttranscriptional regulation of MUC4 by miR-150. HPAF, Panc10.05 and Colo357 PC cells treated with different concentration of miR-150 or non-targeting control (miR-NC) mimic for 48 h. Mock-transfected cells represent cells treated with Lipofectamine 2000 alone. Expression of MUC4 was examined at mRNA (C) and protein (D) levels by quantitative reverse transcription–PCR and western blot analyses, respectively. GAPDH (for RNA) and β-actin (for protein) were used as internal controls. Amplified products from one of replicate wells of MUC4 and GAPDH quantitative PCR were also run on 1% agarose gel (C). Intensities of the immunoreactive bands in western blots were quantified by densitometry (D). Bars represent relative MUC4 expression after normalization with the relative internal control ± SD, *P < 0.05.