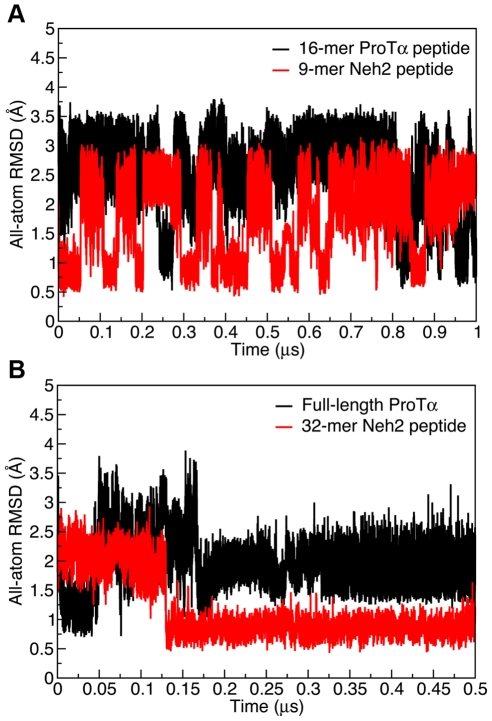
Figure 2. All-atom RMSD values between the MD and crystal structures.
The RMSD values were computed by subtracting the all-atom distance matrix at time t of the MD trajectories from the reference distance matrix determined from the crystal structures of the ProTα and Neh2 peptides bound to Keap1 (PDB ids: 2Z32 and 1X2R respectively) [50], [55]. The distance matrices consisted of residues i through i+3 of the β-turn regions of the ProTα and Neh2 peptides determined from the crystal structures [50], [55].