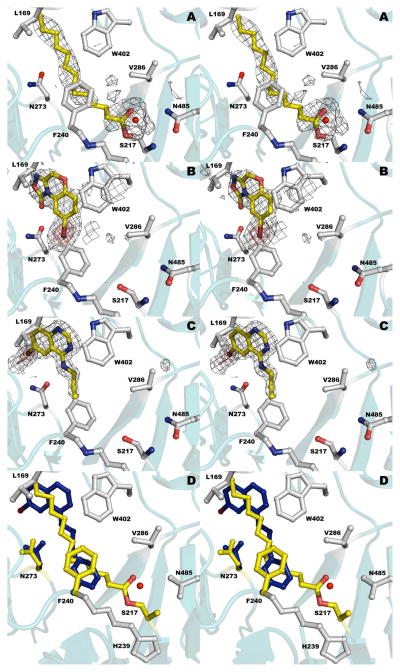
Figure 5.
The liganded catalytic active site of PvdQ. (a) Structure of PvdQ bound to the myristate group derived from incubation of the enzyme with PVDIq. (b) Structure of PvdQ bound to NS2028. (c) Structure of PvdQ bound to SMER28. (d) Superposition of myristoylated enzyme with SMER28. Protein residues are shown in grey and yellow for acylated enzyme and in blue for side chains from the SMER28 structure. In panels a-c, the electron density is calculated with coefficients of the form Fo-Fc generated prior to the inclusion of ligands in the active site. The density is contoured at 2.5σ (grey) and at 8σ (red) in panels b and c.