Abstract
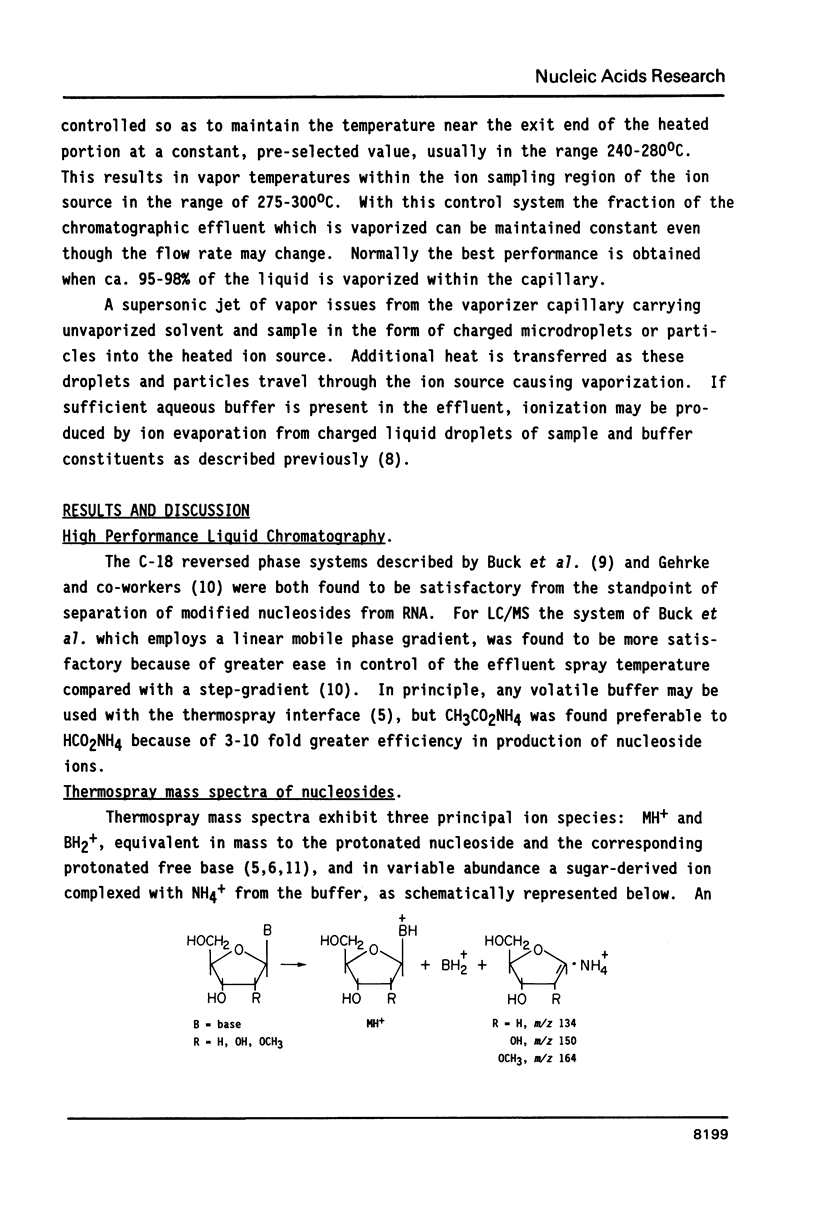
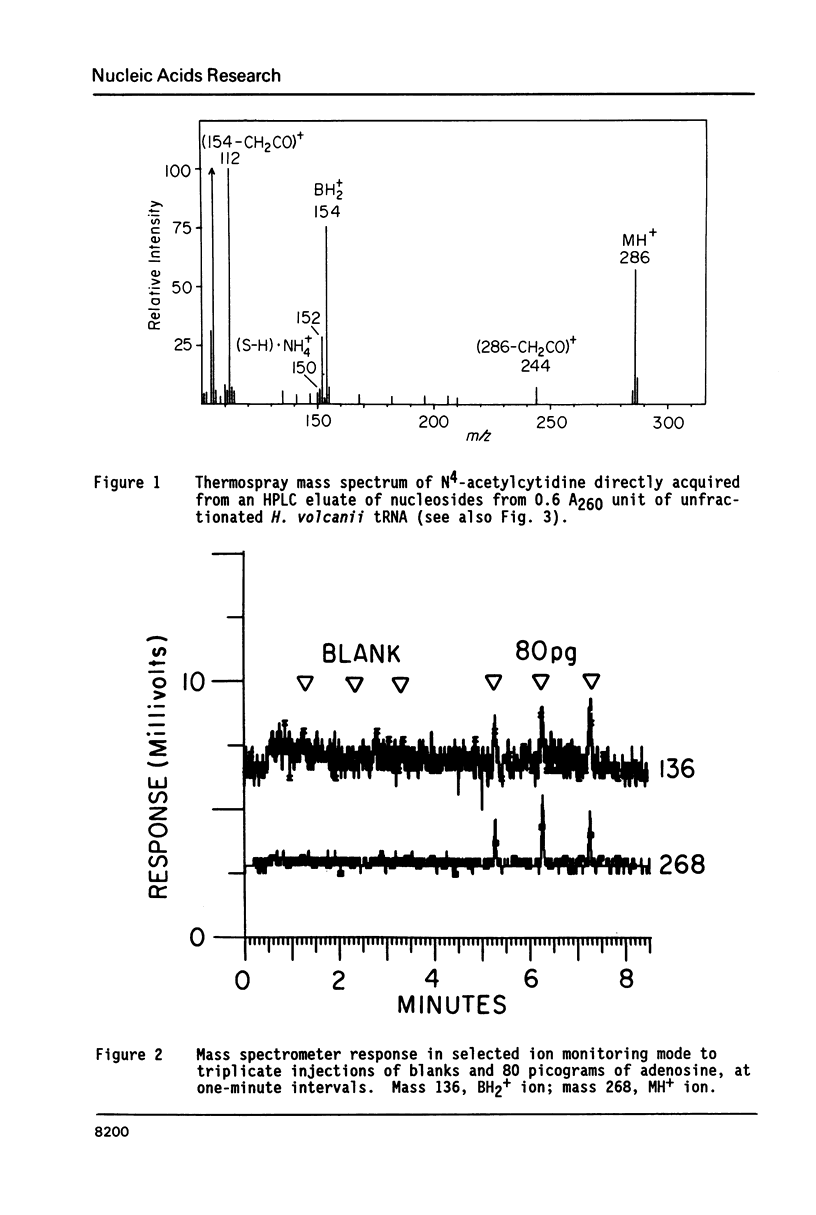
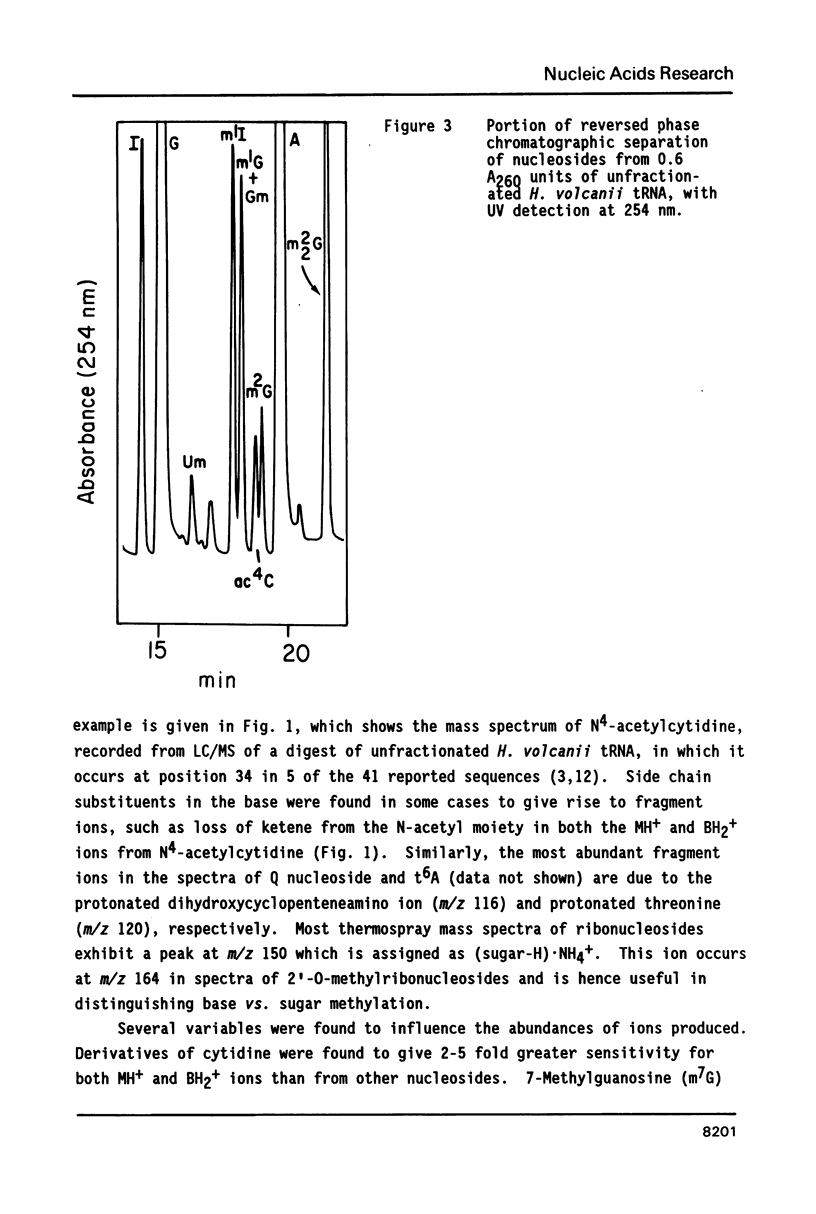
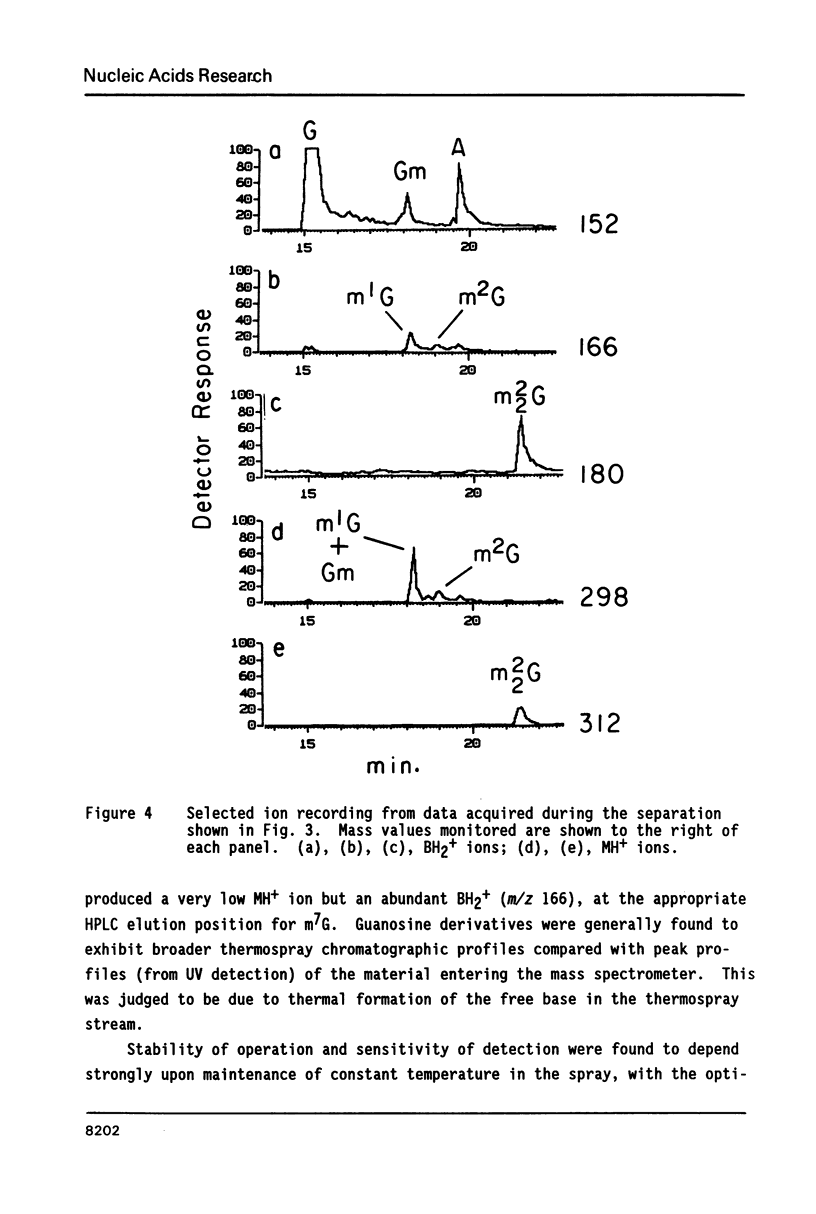
Nucleosides dissolved in aqueous buffered solutions undergo ionization during direct introduction of the solution into a mass spectrometer using a thermospray interface. The principal ions formed represent the protonated molecule, the corresponding protonated free base, and sugar. In addition to potential utility for characterization of new nucleosides, the technique can be used to monitor nucleosides separated from enzymatic hydrolysates by liquid chromatography. The selectivity of chromatographic detection is significantly greater than with UV absorbance alone so that independent detection of components of unresolved chromatographic peaks is usually possible. Detection limits, with signal/noise greater than 10 for most nucleosides, are approximately 0.1-1 ng per component for selected ion monitoring and 10-50 ng for full-scan mass spectra. Examples are given from the detection of modified nucleosides in enzymatic hydrolysates of 0.05 A260 units (2.5 micrograms) of rabbit liver tRNAVal and of unfractionated H. volcanii tRNA.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buck M., Connick M., Ames B. N. Complete analysis of tRNA-modified nucleosides by high-performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal Biochem. 1983 Feb 15;129(1):1–13. doi: 10.1016/0003-2697(83)90044-1. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C., Zumwalt R. W. Chromatography of nucleosides. J Chromatogr. 1980 Jan 25;188(1):129–147. doi: 10.1016/s0021-9673(00)88424-1. [DOI] [PubMed] [Google Scholar]
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984 Aug 10;259(15):9461–9471. [PubMed] [Google Scholar]
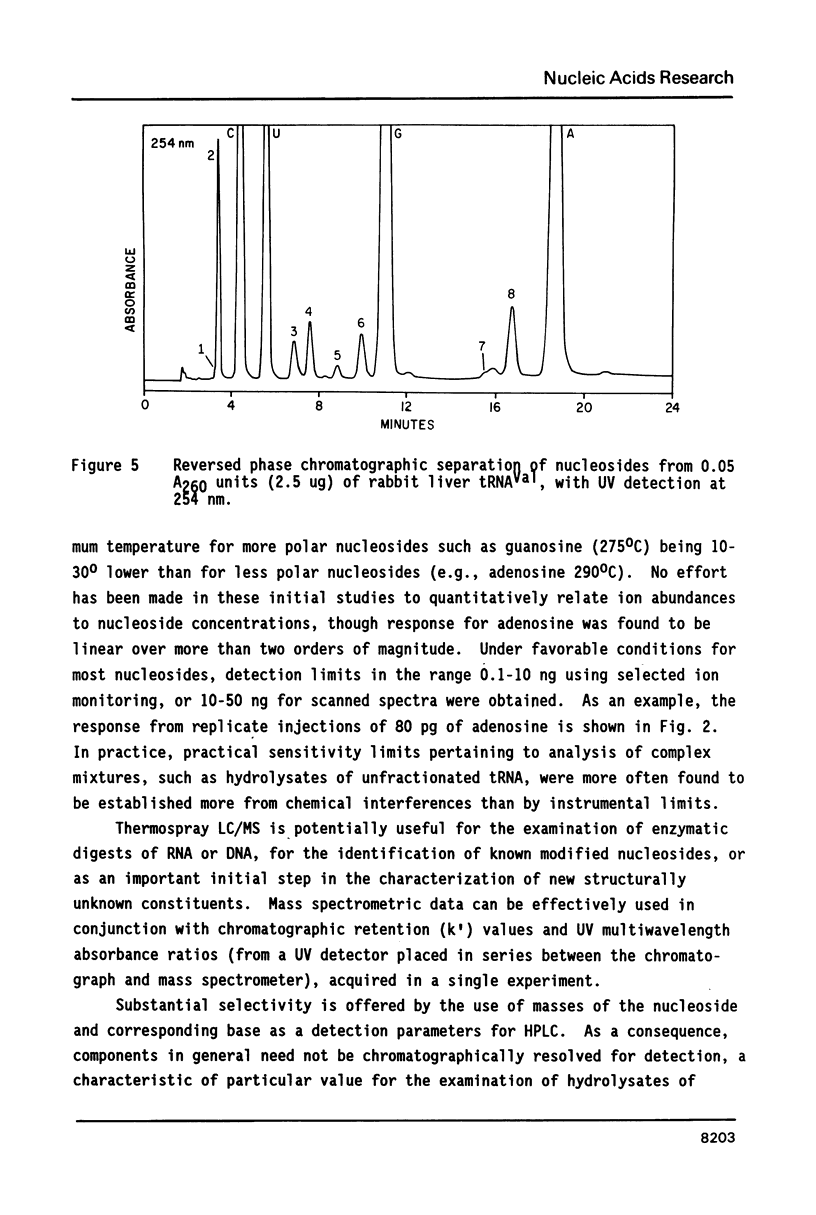
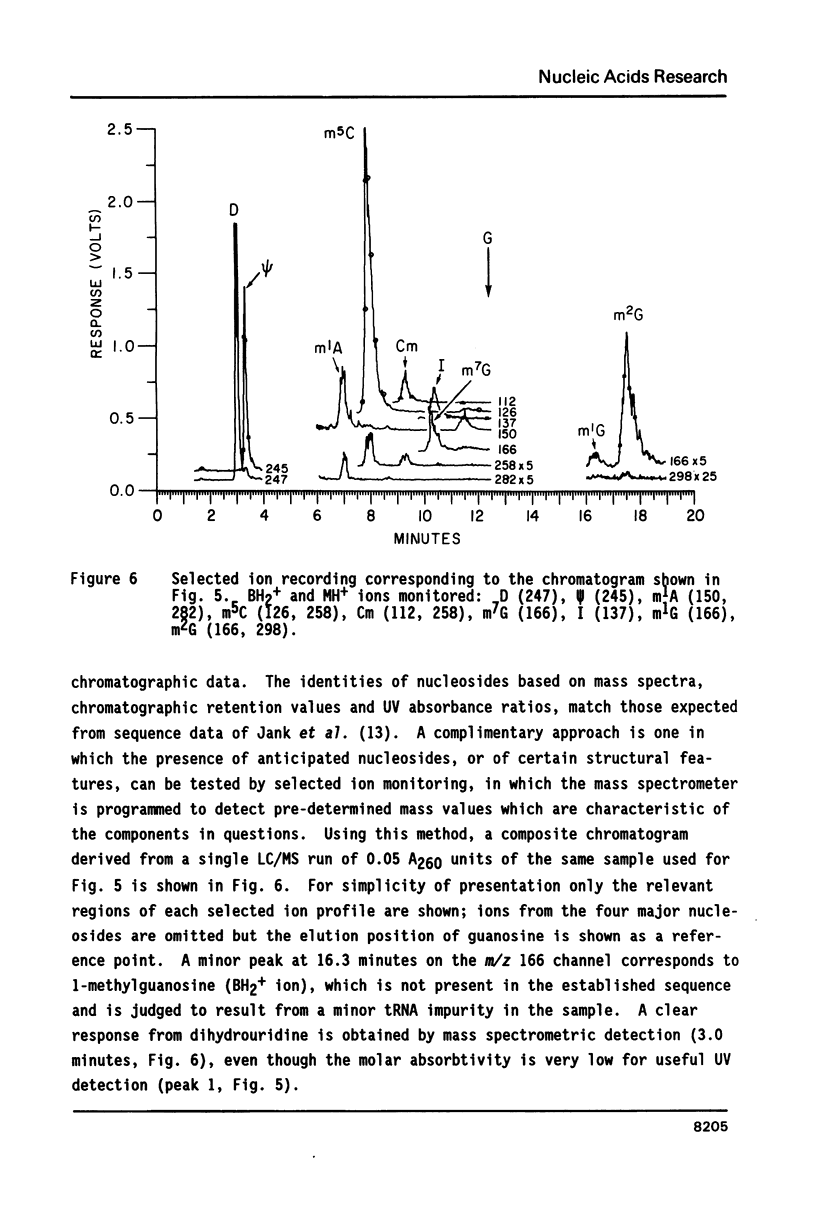
- Jank P., Shindo-Okada N., Nishimura S., Gross H. J. Rabbit liver tRNA1Val:I. Primary structure and unusual codon recognition. Nucleic Acids Res. 1977 Jun;4(6):1999–2008. doi: 10.1093/nar/4.6.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Moll J., Meissner F., Hartmann T. Compilation of tRNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r1–49. doi: 10.1093/nar/13.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. A. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]


