Introduction
Genetic studies of bone mass, bone mineral content (BMC), and/or bone mineral density (BMD) have demonstrated these measures to be highly heritable [1–11]. As substantial evidence for an additive genetic contribution to adult bone mass and risk for osteoporosis has now been well established, genetic research in this area has moved increasingly toward the identification of genomic regions or candidate genes influencing variation in such traits [12–14]. Recent genetic linkage and genome-wide association studies of bone mass in adults have identified regions on at least 11 separate chromosomes [7,15–18] potentially harboring genes that play a role in regional or total body BMD [12,18–25].
It is evident that researchers are closing in on specific genes influencing the maintenance or loss of adulthood bone mass. Research into childhood skeletal genetics, however, is less abundant and generally focuses on genes underlying specific bone disorders. This paucity of information regarding the genetics of pediatric skeletal growth is unfortunate given that the health of a child’s skeleton, and the attainment of peak bone mass in young adulthood, is vital to the health of the adult skeleton. Further, the genetic underpinnings of bone accrual during childhood may differ from that of bone maintenance and/or loss during adulthood. This paper presents the results of genome-wide linkage analyses of measures of childhood cortical bone health.
Materials and methods
Study Sample
The study sample is comprised of a subset of 600 individuals participating in the Fels Longitudinal Study, the world’s longest-running longitudinal study of human growth, development, and body composition change over the life span [26]. Subjects are primarily Caucasian and live (or lived at the time of evaluation) in southwest Ohio or immediately surrounding regions. Children represented in this sample are considered normal and healthy and were not selected for any disease or bone related trait. Specifically, we examined hand-wrist radiographs of these 600 individuals (317 boys, 283 girls) taken when they each were 10 years old. The majority of children were within 2 weeks of their 10th birthday at the time of radiography. It is important to emphasize that although individuals in this study span up to three generations (see below), all individuals were sampled at the same chronological age. This single age was chosen in order to minimize variation in bone measurements that could be attributed to age or growth status, and to minimize sex differences that normally arise with puberty. In doing so, we maximize the ability to detect genetic involvement in the study traits. Boys and girls were analyzed together in all genetic analyses (all bone measures were shown to have no significant differences between the sexes; Duren et al.[9]).
Because of the longevity of the Fels Longitudinal Study and the participation of individuals from several dozen large nuclear and extended families, the study sample of 600 predominantly white individuals from 144 families contains many relative pairings spanning up to three generations. Median family size for this sample is 4, the mean family size is 6, and the maximum number of individuals examined in a single family is 59. There are 2,925 relative pairings among the 600 individuals in the study sample. Pairings range from 1st degree relatives (e.g., sibling and parent-offspring pairs) to 8th degree relatives (e.g., 3rd cousins twice removed).
Bone Measurements
Radiographic measurements of a cylindrical bone such as the second metacarpal are generally more reliable than those taken from less cylindrical bones [27]. Because bone tissue is relatively evenly distributed around the circumference in the second metacarpal, slight variation in radiographic positioning does not have a large effect on metrics. Additionally, measures of cortical bone are less affected by short-term temporal fluctuation in bone mass caused by seasonal variation and weight changes than measures of BMD or BMC from dual energy x-ray absorptiometry (DEXA) [28].
Quantitative measurements of bone mass from radiographs were obtained from the second metacarpal in each individual according to standard methods [29]. Bone diameter, medial cortical thickness, lateral cortical thickness, and medullary diameter were each measured in millimeters on the second metacarpal (see Duren et al.[9]). All measurements were taken directly from original radiographs using digital Mitutoyo calipers with direct computer input. Two additional variables were derived from these measures: 1) total cortical thickness, calculated as the sum of medial and lateral cortex; and 2) cortical index (CI), calculated as total cortex/bone diameter [after 30]. Sample-wide coefficients of variation ranged from 11.8 to 15.5% [9].
Genotyping and the Whole Genome Linkage Map
Whole genome marker genotyping
Of the 600 individuals sampled in this study, 440 were genotyped for ~400 highly polymorphic genetic markers on autosomes using the ABI PRISM Linkage Mapping Set Version 2 (Applied Biosystems, Foster City, CA). This mapping set consists of fluorescently labeled PCR primers that amplify dinucleotide repeat microsatellite loci (STRs) selected from the Genethon human linkage map[31–33]. The ABI PRISM Linkage Mapping Set-MD10 consists of 400 markers. The set is designed to create a map with markers spaced on average 10cM apart (range 2.4 to 24.1 cM).
Genotyping error checking
Genotypes assigned to individuals were checked for Mendelian consistency using the PEDSYS[34] program INFER. Discrepancies were initially referred to the lab for re-typing of markers in order to resolve them. If discrepancies could not be resolved by re-typing, the program SimWalk2 [35] was used to make decisions about the appropriate genotypes to exclude in that situation.
Estimation of allele frequencies
Estimates of allele frequencies for each genetic marker locus were obtained using Sequential Oligogenic Linkage Analysis Routines (SOLAR) [36]. The linkage map was based on the deCODE map with non-deCODE markers added by interpolation using physical location.
Statistical and Genetic Analyses
We used a maximum likelihood-based variance decomposition approach implemented in SOLAR [36] to estimate heritability for each skeletal variable and to conduct multipoint linkage analysis of each skeletal trait. Univariate linkage analyses were performed to locate areas of the genome that harbor genes (quantitative trait loci, QTL) that influence measures of second metacarpal morphology. To examine QTL linked to our skeletal traits, the covariance matrix (Ω) for a pedigree is:
where σ2q is the genetic variance in the trait due to the QTL, Π is a matrix whose elements provide the number of genes that a relative pair share IBD at that QTL that is linked to a genetic marker locus, σ2a is the genetic variance due to additive genetic factors, Φ is the kinship matrix, σ2e is the variance due to individual-specific environmental effects, and I is an identity matrix.
The hypothesis of linkage was tested by comparing the likelihood of a restricted model in which σ2q (the genetic variance due to the QTL) equals zero (no linkage) to a model in which σ2q is estimated. The difference between these two log10 likelihoods produces a LOD (log-odds) score that is the equivalent of the classical LOD score of linkage analysis [38].
To control for the genome-wide false positive rate, we calculated genome-wide p-values for each LOD score using a modification of a method suggested by Feingold et al. [39] that takes into account pedigree complexity and the finite marker density of the linkage map. Accordingly, our threshold for significant evidence of linkage (corresponding to genome-wide α = 0.05) was LOD = 2.87, while suggestive evidence of linkage occurred at LOD=1.67. These values correspond to the expected false positive rates of once per 20 (“significant”) and once per 10 (“suggestive”) genome-wide linkage screens [40].
The power to detect genetic effects was improved by accounting for environmental contributions to the phenotypic variance. Prior to all analyses, we used likelihood ratio tests to screen each of the following covariates for significant mean effects on the second metacarpal cortical variables: age, sex, age2, age × sex, age2 × sex, and skeletal age.
Results
Basic quantitative genetic analysis using the complete data set of 600 phenotyped 10-year olds revealed significant (p<0.001) additive genetic components to the total variance for all skeletal variables examined. Heritability (h2) estimates ranged from 0.45 ±0.11 (for lateral cortical thickness) to 0.71 ±0.10 (for cortical index). Details and discussion of these results have been published previously [9]. In addition to these variables measured at the narrowest location on the metacarpal shaft, cortical index was also recorded and analyzed at the midshaft location of the metaphysis (heritability estimate for this trait was 0.61 ±0.11).
Whole genome linkage analyses in the sub-sample of 440 genotyped 10-year-olds revealed several chromosomal regions harboring genes influencing childhood skeletal variation. Significant QTL (LOD > 2.87) were identified for three of the bone traits (see Table 1).
Table 1.
Quantitative Trait Loci for childhood bone traits in Fels Longitudinal Study participantsa.
| Trait | Cytogenic location | cMb | LOD | #BP in 1-LOD interval | # Genes in 1-LOD interval | ||
|---|---|---|---|---|---|---|---|
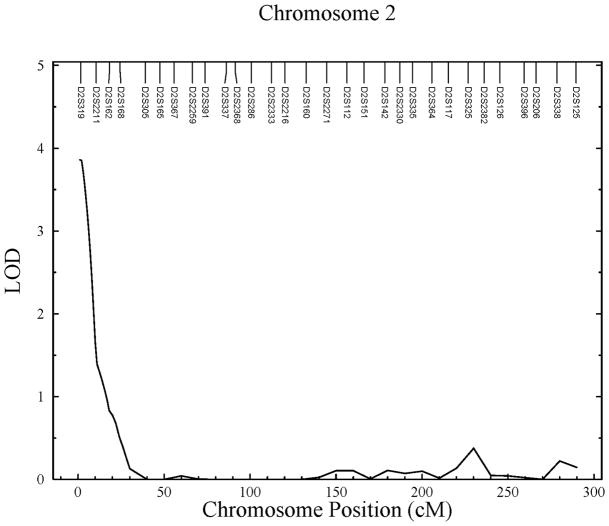
| Medial Cortical Thickness | 2p25.2 | 1 | 3.86 | 0.58 | 6,607,526 | 18 | |
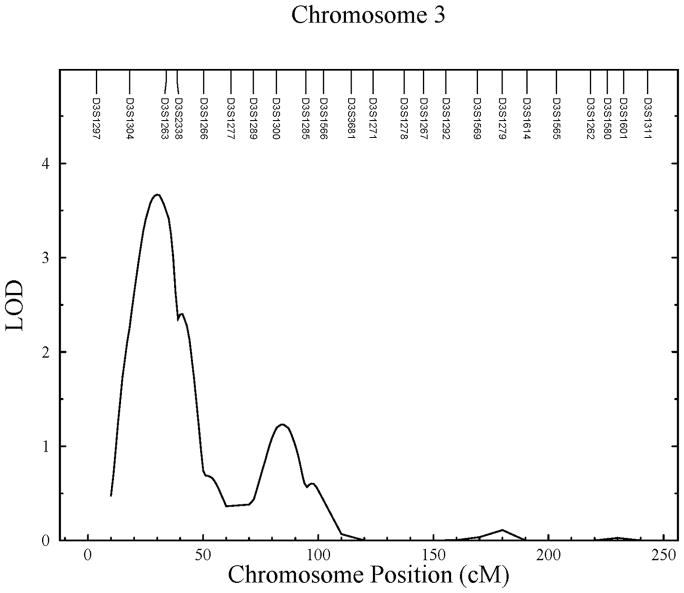
| Lateral Cortical Thickness | 3p25.3 | 30 | 3.67 | 0.54 | 18,944,086 | 99 | |
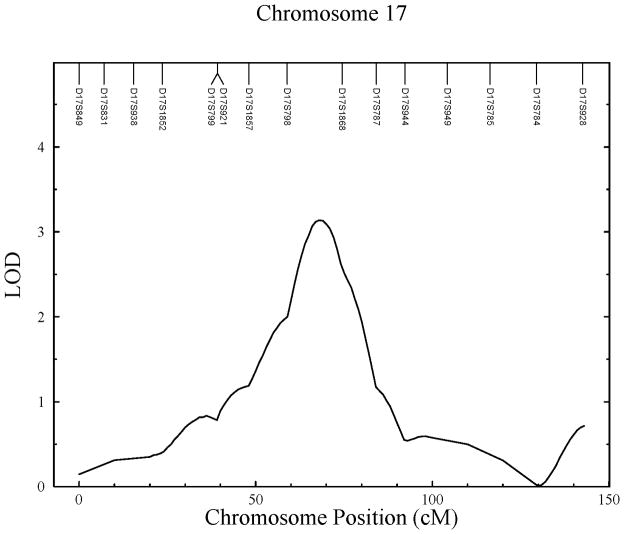
| Cortical Index | 17q21.2 | 68 | 3.14 | 0.55 | 18,629,864 | 392 |
Maximum likelihood heritability estimates from linkage models: is the QTL-specific heritability, i.e., the proportion of the residual phenotypic variance due to the effect of the QTL.
Location in centimorgans (cM) from pter-most marker locus in the human genetic linkage map for that chromosome.
Significant linkages were identified on chromosomes 2p, 3p, and 17q for the traits medial cortex, lateral cortex, and cortical index, respectively (see Figure 1). The location of the linkage peak for medial cortical thickness (LOD = 3.86) is near marker D2S319, which corresponds to chromosomal region 2p25.2. The 1-LOD interval of this linkage spans an area 6.6Mb wide starting at the pter end of the chromosome. This QTL was estimated to account for 58% of the detected residual phenotypic variance in second metacarpal medial cortical thickness.
Figure 1.
Linkage results for childhood cortical bone traits. A) Significant linkage found on chromosome 2 for medial cortical thickness; B) significant linkage found on chromosome 3 for lateral cortical thickness; and C) significant linkage found on chromosome 17 for cortical index.
The QTL for lateral cortical thickness (LOD = 3.67) is located between markers D3S1304 and D3S1263, overlapping chromosomal regions 3p26.1 and 3p25.3. This linkage has an 18.9Mb wide 1-LOD interval, and accounts for 45% of the detected residual phenotypic variance. A second significant linkage (LOD = 3.30) was found at the same region for total cortical thickness which is directly related to lateral cortical thickness. Suggestive linkage (LOD = 2.27) was also found to this region for medial cortical thickness.
The third significant QTL identified in this analysis is for cortical index at metaphyseal mid-shaft (LOD = 3.14). This QTL was found at chromosomal region 17q21.2 region, between markers D17S798 and D17S1868. This linkage has a 1-LOD interval spanning 18.6Mb and was estimated to account for 55% of the detected residual phenotypic variance. Additional suggestive linkage (LOD = 2.11) was found to this region for medial cortical thickness.
Discussion
The current investigation represents the first genetic linkage study focusing on skeletal mass/cortical thickness phenotypes in young children. The field of skeletal biology/bone health has been very successful, notably in recent years, in identifying genomic regions harboring genes responsible for bone maintenance and loss. Linkage studies of bone mass phenotypes have focused mainly on adults and groups at risk for osteoporosis. However, the genetic underpinnings of childhood bone health may have significant impact on adult bone health. Here, we found cortical bone thickness in 10-year-old children to be significantly linked to three genomic regions. Each of these regions contains plausible candidate genes, some of which have previously been identified by genetic studies of adult bone mass (see below). The skeletal phenotypes in our study are based on radiographic representations of bone. Although most other genetic studies measure bone mass by DEXA, we are encouraged that our analyses have confirmed areas initially identified in studies of adults using different methodologies. Further, our analyses have additionally identified new areas of the genome influencing the skeletal system during childhood.
It is generally accepted that genes regulating bone metabolism may influence regional bone mass in different ways, and may represent only a part of the genetic regulation of “whole body” systemic bone mass [16,41]. In fact, most investigations of regional bone density or morphological parameters (e.g., hip geometry) typically identify different genomic regions influencing the various bone traits under investigation. While the pleiotropic effects of some genes will influence global aspects of bone remodeling, it is clear that there are site-specific genetic effects on bone. This may have much to do with the functional environment of the skeletal element or trait, so that a bone or region of a bone under substantial mechanical stress (e.g., during locomotion) may be differentially influenced by genes than an element free of that type of mechanical strain. In addition to regional differences in genetic control, it is reasonable that the genetic “landscape” changes over the course of one’s life. For example, genes that influence bone accrual in childhood are likely different than those influencing bone resorption and remodeling in adulthood, which may also differ from those influencing mineral maintenance through bone metabolism [42].
Candidate Genes
The three significant QTL identified for bone health phenotypes in this sample of 10-year-old children correspond to genomic regions identified by other groups in samples of adult populations. Furthermore, the 1-LOD intervals surrounding each of our linkage peaks contain plausible positional candidate genes relevant to skeletal health, and importantly, many of these gene products are vital to the accrual of bone mass.
Genomic Region 2p25.2
Areas of the 2p region have been identified by other studies as harboring genomic “hot spots” for bone-related traits [e.g., 20,22,43,44,45,46,47,48,49]. Our linkage at 2p25.2 lies in close proximity to the region that has previously shown linkage to adult spinal BMD [7,22]. This region contains several genes previously recognized as candidate genes involved in adult skeletal health, including calmodulin 2 (CALM2), serine/theronine (STK), and pro-opiomelanocortin (POMC)[22]. The CALM2 gene has specific roles in bone development and is involved in control of osteoblast proliferation and differentiation, and thus may have its adulthood effects on bone rooted in early life bone development. The STK gene, also located in this region, is expressed by cancellous osteoblasts and has been reported by at least two other studies of skeletal genetics as a candidate gene for bone mass [20,22]. Additionally, this region of chromosome 2p houses the POMC gene, involved in calcium homeostasis, which has also been identified as a candidate gene for bone health in perimenopausal women and similarly aged men [46]. Our evidence of linkage to this region confirms the potential role of these candidate genes influencing not only bone metabolism throughout life, but also their influential role in childhood bone growth and apposition.
An additional positional candidate gene that lies at the peak of our QTL in this region is the thyroid peroxidase (TPO) gene. TPO encodes for a membrane-bound glycoprotein that plays an essential role in thyroid gland function. While it has not been identified as a candidate gene in previous linkage or association studies of bone health in adults, the TPO gene nevertheless is an attractive positional candidate gene for childhood bone accrual in the chromosomal region 2p25.2. Thyroid hormones are essential for bone growth, metabolism, and attainment of peak bone mass [50,51]. Mutations in TPO are associated with thyroid disorders (e.g., congenital goiter and hypothyroidism)[52], and thus likely influence bone metabolism. Further work is needed to confirm any role the TPO gene may have on pediatric skeletal health as measured in the current study.
Genomic Region 3p25.3
The 3p25.3 region identified by our linkage analysis is consistent with results of other genetic studies of adult skeletal phenotypes [53–56]. In each of these studies of bone mass, lumbar spine/spine BMD has shown linkage to the 3p25 region. Additionally, bone density of the ultradistal forearm showed linkage to the 3p25 region [55]. While none of the linkages identified in these studies reached genome-wide significance, they do identify this region as one of potential interest. The 1-LOD support interval on chromosome 3p (3p26.1 - 3p25.3) in our study contains genes of potential interest to child bone health including the calcium/calmodulin-dependent protein kinase I (CaMK1) gene. CaMK1 is expressed in many tissues, including osteoblasts of developing long bones [57]. Because calcium/calmodulin directly activates the CaMK1 by binding to the enzyme, and indirectly promotes phosphorylation and synergistic activation of the enzyme, the CaMK1 gene, though ubiquitous throughout the body, may have an indirect impact on bone metabolism [57,58].
One candidate gene within the 1-LOD interval of our linkage results that has not been identified in other analyses is the oxytocin receptor gene (OXTR). OXTR is a member of the G protein-coupled receptor superfamily [59]. Oxytocin is classically known for its role in smooth muscle contraction in the uterus and mammary tissue [60], but evidence suggests OXTR is also an important bone mass regulator. In 1999 the receptor itself (OTR) was found to be functional in osteoblasts, mediating increases in intracellular calcium levels as well as prostaglandin E2 synthesis [61]. In 2002, the OTR was discovered to be functional in osteoclasts as well, increasing proliferation of pre-osteoclasts upon ligand binding [62]. This finding was confirmed by Petersson [63] who also showed that oxytocin is important for bone metabolism, stimulating interleukin (IL)-6 production by osteoblasts. Oxytocin itself is also involved in skeletal health. A recent study of post-menopausal women showed an association between low levels of circulating oxytocin and severe osteoporosis [64]. Given these discoveries, and the delicate balance of osteoclastic and osteoblastic activity in bone remodeling, it would appear that oxytocin and its receptor may be important for bone mass regulation, especially during childhood.
To date, we are unaware of any other genetic study that has found significant linkage of bone mass traits to OXTR. Because of oxytocin receptor’s role in bone regulation, the OXTR gene is an excellent candidate gene for bone mass, especially during childhood. While OXTR has not been identified by other genetic linkage studies, it is important to note that the OTR has binding affinity not only to oxytocin, but also to estrogen [61]. Estrogen has long been known to influence the regulation of bone mass, and several genetic studies have identified estrogen and estrogen-related genes responsible for bone mass variation [65].
Genomic Region 17q21.2
The significant linkage found on chromosome 17 for cortical index corresponds to a region containing the sclerostin (SOST) gene. Sclerostin is a protein expressed nearly exclusively in osteocytes [66,67], which inhibits portions of the Wnt-signaling pathway, thereby inhibiting bone formation. Mutations in the gene that prevent sclerostin production result in sclerosteosis, a condition characterized by high bone mass throughout the body [68]. While those affected by this autosomal recessive trait exhibit marked increased bone mass, heterozygous carriers also exhibit higher than normal BMD values. Given the role of sclerostin in bone health, the SOST gene is likely to have a large role in bone acquisition and remodeling during the early and mid-childhood periods.
Conclusions
Replication of chromosomal regions has become an important criterion for genetic studies. Although we are unaware of any other study of child bone health that has reported linkage to any of these regions, we have provided evidence of significant genetic linkage to regions identified by adult studies as well as regions known to harbor genes influencing variation in bone morphology. We are also analyzing these bone traits at other childhood ages to determine if these regions are specific to bone health at a single time point or if the genes in these regions are influential on bone throughout childhood and/or adulthood.
The identification of three genomic regions influencing childhood bone health is a significant step for the field. Confirmation of the role of the specific genes contained in these regions may ultimately lead to better medical care for children with disorders of skeletal mass, and may shed light on gene action during childhood that has a lasting impact on skeletal health throughout the life span.
Highlights.
Cortical bone accrual in childhood is under significant genetic control.
Childhood cortical bone is linked to genes on chromosomes 2p, 3p and 17q.
Candidate genes for childhood bone health include the sclerostin and oxytocin receptor genes.
Acknowledgments
The authors wish to thank the editors and two anonymous reviewers for helpful comments. We would like to express our gratitude to the staff at the Lifespan Health Research Center for assistance in data collection. We also thank the dedicated participants of the Fels Longitudinal Study for their contribution to science.
This work was supported by grants from Wright State University Boonshoft School of Medicine, and the National Institutes of Health (R01HD05247, R01AR055927, R01HD036342, R01HD012252, and R37MH59490).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Christian JC, Yu PL, Slemenda CW, Johnston CC., Jr Heritability of bone mass: a longitudinal study in aging male twins. Am J Hum Genet. 1989;44:429–433. [PMC free article] [PubMed] [Google Scholar]
- 2.Gueguen R, Jouanny P, Guillemin F, Kuntz C, Pourel J, Siest G. Segregation analysis and variance components analysis of bone mineral density in healthy families. J Bone Miner Res. 1995;10:2017–2022. doi: 10.1002/jbmr.5650101223. [DOI] [PubMed] [Google Scholar]
- 3.Kammerer CM, Sparks ML, Rogers J. Effects of age, sex, and heredity on measures of bone mass in baboons (Papio hamadryas) J Med Primatol. 1995;24:236–242. doi: 10.1111/j.1600-0684.1995.tb00176.x. [DOI] [PubMed] [Google Scholar]
- 4.Hopper JL, Green RM, Nowson CA, Young D, Sherwin AJ, Kaymakci B, Larkins RG, Wark JD. Genetic, common environment, and individual specific components of variance for bone mineral density in 10- to 26-year-old females: a twin study. Am J Epidemiol. 1998;147:17–29. doi: 10.1093/oxfordjournals.aje.a009361. [DOI] [PubMed] [Google Scholar]
- 5.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocrine Reviews. 2002;23:303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 6.Livshits G, Yakovenko K, Kobyliansky E. Quantitative genetic study of radiographic hand bone size and geometry. Bone. 2003;32:191–198. doi: 10.1016/s8756-3282(02)00954-7. [DOI] [PubMed] [Google Scholar]
- 7.Wynne F, Drummond FJ, Daly M, Brown M, Shanahan F, Molloy MG, Quane KA. Suggestive linkage of 2p22–25 and 11q12–13 with low bone mineral density at the lumbar spine in the irish population. Calcif Tissue Int. 2003;72:651–658. doi: 10.1007/s00223-002-2086-2. [DOI] [PubMed] [Google Scholar]
- 8.Econs MJ, Koller DL, Hui SL, Fishburn T, Conneally PM, Johnston CC, Jr, Peacock M, Foroud TM. Confirmation of linkage to chromosome 1q for peak vertebral bone mineral density in premenopausal white women. Am J Hum Genet. 2004;74:223–228. doi: 10.1086/381401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duren DL, Sherwood RJ, Choh AC, Czerwinski SA, Cameron CW, Lee M, Sun SS, Demerath EW, Siervogel RM, Towne B. Quantitative genetics of cortical bone mass in healthy 10-year-old children from the Fels Longitudinal Study. Bone. 2007;40:464–470. doi: 10.1016/j.bone.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havill LM, Allen MR, Bredbenner TL, Burr DB, Nicolella DP, Turner CH, Warren DM, Mahaney MC. Heritability of lumbar trabecular bone mechanical properties in baboons. Bone. 2010;46:835–840. doi: 10.1016/j.bone.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, Song YM, Sung J, Lee K, Kim YS, Park YS. Genetic influence on bone mineral density in Korean twins and families: the healthy twin study. Osteoporos Int. 2011 doi: 10.1007/s00198-011-1685-z. [DOI] [PubMed] [Google Scholar]
- 12.Ralston SH. Science, medicine, and the future: Osteoporosis. Br Med J. 1997;315:469–472. [Google Scholar]
- 13.Liu Y-Z, Liu Y-J, Recker RR, Deng H-W. Molecular studies of identification of genes for osteoporosis: the 2002 update. Journal of Endocrinology. 2003;177:147–196. doi: 10.1677/joe.0.1770147. [DOI] [PubMed] [Google Scholar]
- 14.Rubin LA, Patel MS, Cole DE. Genetic determinants of bone mass acquistion and risk for osteoporosis. Drug Dev Res. 2000;49:216–226. [Google Scholar]
- 15.Karasik D, Cupples LA, Hannan MT, Kiel DP. Age, gender, and body mass effects on quantitative trait loci for bone mineral density: the framingham study. Bone. 2003;33:308–316. doi: 10.1016/s8756-3282(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 16.Wilson SG, Reed PW, Bansal A, Chiano M, Lindersson M, Langdown M, Prince RL, Thompson D, Thompson E, Bailey M, Kleyn PW, Sambrook P, Shi MM, Spector TD. Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. American Journal of Human Genetics. 2003;72:144–155. doi: 10.1086/345819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, Zhang YY, Long JR, Xu FH, Liu YZ, Xiao P, Zhao LJ, Xiong DH, Liu YJ, Dvornyk V, Rocha-Sanchez S, Liu PY, Li JL, Conway T, Davies KM, Recker RR, Deng HW. A genome-wide linkage scan for bone mineral density in an extended sample: evidence for linkage on 11q23 and Xq27. J Med Genet. 2004;41:743–751. doi: 10.1136/jmg.2004.020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li GH, Cheung CL, Xiao SM, Lau KS, Gao Y, Bow CH, Huang QY, Sham PC, Kung AW. Identification of QTL genes for BMD variation using both linkage and gene-based association approaches. Hum Genet. 2011 doi: 10.1007/s00439-011-0972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison NA, Qi JC, Tokita A, Kelly PJ, Crofts L, Nguyen TV, Sambrook PN, Eisman JA. Prediction of bone density from vitamin D receptor alleles. Nature. 1994;367:284–287. doi: 10.1038/367284a0. [DOI] [PubMed] [Google Scholar]
- 20.Devoto M, Shimoya K, Caminis J, Ott J, Tenenhouse A, Whyte MP, Sereda L, Hall S, Considine E, Williams CJ, Tromp G, Kuivaniemi H, Ala-Kokko L, Prockop DJ, Spotila LD. First-stage autosomal genome screen in extended pedigrees suggests genes predispsing to low bone mineral density on chromosomes 1p, 2p and 4q. European Journal of Human Genetics. 1998;6:151–157. doi: 10.1038/sj.ejhg.5200169. [DOI] [PubMed] [Google Scholar]
- 21.Devoto M, Specchia C, Li H-H, Caminis J, Tenenhouse A, Rodriguez H, Spotila LD. Variance component linkage analysis indicates a QTL for femoral neck bone mineral density on chromosome 1p36. Human Molecular Genetics. 2001;10:2447–2452. doi: 10.1093/hmg/10.21.2447. [DOI] [PubMed] [Google Scholar]
- 22.Niu T, Chen C, Cordell H, Yang J, Wang B, Wang Z, Fang Z, Schork NJ, Rosen CJ, Xu X. A genome-wide scan for loci linked to forearm bone mineral density. Hum Genet. 1999;104:226–233. doi: 10.1007/s004390050940. [DOI] [PubMed] [Google Scholar]
- 23.Carn G, Koller DL, Peacock M, Hui SL, Evans WE, Conneally PM, Johnston CC, Jr, Foroud T, Econs MJ. Sibling pair linkage and association studies between peak bone mineral density and the gene locus for the osteoclast-specific subunit (OC116) of the vacuolar proton pump on chromosome 11p12–13. J Clin Endocrinol Metab. 2002;87:3819–3824. doi: 10.1210/jcem.87.8.8740. [DOI] [PubMed] [Google Scholar]
- 24.Koller DL, White KE, Liu G, Hui SL, Conneally PM, Johnston CC, Econs MJ, Foroud T, Peacock M. Linkage of structure at the proximal femur to chromosomes 3, 7, 8, and 19. J Bone Min Res. 2003;18:1057–1065. doi: 10.1359/jbmr.2003.18.6.1057. [DOI] [PubMed] [Google Scholar]
- 25.Koller DL, Econs MJ, Morin PA, Christian JC, Hui SL, Parry P, Curran ME, Rodriguez LA, Conneally PM, Joslyn G, Peacock M, Johnston CC, Foroud T. Genome screen for QTLs contributing to normal variation in bone mineral density and osteoporosis. J Clin Endocrinol Metab. 2000;85:3116–3120. doi: 10.1210/jcem.85.9.6778. [DOI] [PubMed] [Google Scholar]
- 26.Roche AF. Growth, Maturation, and Body Composition: The Fels Longitudinal Study. Cambridge University Press; Cambridge: 1992. [Google Scholar]
- 27.Garn SM. The Earlier Gain and the Later Loss of Cortical Bone. Bannerstone House; Springfield, IL: 1970. [Google Scholar]
- 28.Karasik D, Ginsburg E, Livshits G, Pavlovsky O, Kobyliansky E. Evidence of major gene control over cortical bone loss in humans. Genet Epidemiol. 2000;19:410–421. doi: 10.1002/1098-2272(200012)19:4<410::AID-GEPI11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 29.Plato CC, Fox KM, Tobin JD. Skeletal Changes in Human Aging. In: Crews DE, Garruto RM, editors. Biological Anthropology and Aging: Perspectives on human variation over the life span. Oxford University Press; New York: 1994. [Google Scholar]
- 30.SINCLAIR RJG, KITCHEN AH, TURNER RWD. THE MARFAN SYNDROME. QJM. 1960;29:19–46. [Google Scholar]
- 31.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 32.Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J. The 1993–94 Genethon human genetic linkage map. Nat Genet. 1994;7:246–339. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- 33.Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseix G, Lathrop M. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- 34.Dyke B. Populations Genetics Laboratory Technical Report No 2. 2. Southwest Foundation for Biomedical Research; 1994. PEDSYS, a pedigree data management system user’s manual. [Google Scholar]
- 35.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 36.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott J. Analysis of Human Genetic Linkage. The Johns Hopkins University Press; Baltimore and London: 1988. [Google Scholar]
- 39.Feingold E, Brown PO, Siegmund D. Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am J Hum Genet. 1993;53:234–251. [PMC free article] [PubMed] [Google Scholar]
- 40.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 41.MacGregor AJ, Snieder H, Keen RW, Spector T. Do different sets of genes influence trabecular and cortical bone density at different anatomical sites? J Bone Miner Res. 1997;(Suppl 12):S492. [Google Scholar]
- 42.Lutz J, Tesar R. Mother-daughter pairs: spinal and femoral bone densities and dietary intakes. Am J Clin Nutr. 1990;52:872–877. doi: 10.1093/ajcn/52.5.872. [DOI] [PubMed] [Google Scholar]
- 43.Peacock M, Koller DL, Lai D, Hui S, Foroud T, Econs MJ. Sex-specific quantitative trait loci contribute to normal variation in bone structure at the proximal femur in men. Bone. 2005;37:467–473. doi: 10.1016/j.bone.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peacock M, Koller DL, Fishburn T, Krishnan S, Lai D, Hui S, Johnston CC, Foroud T, Econs MJ. Sex-specific and non-sex-specific quantitative trait loci contribute to normal variation in bone mineral density in men. J Clin Endocrinol Metab. 2005;90:3060–3066. doi: 10.1210/jc.2004-2143. [DOI] [PubMed] [Google Scholar]
- 45.Karasik D, Dupuis J, Cupples LA, Beck TJ, Mahaney MC, Havill LM, Kiel DP, Demissie S. Bivariate linkage study of proximal hip geometry and body size indices: the framingham study. Calcif Tissue Int. 2007;81:162–173. doi: 10.1007/s00223-007-9052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang QY, Kung AW. The association of common polymorphisms in the QPCT gene with bone mineral density in the Chinese population. J Hum Genet. 2007;52:757–762. doi: 10.1007/s10038-007-0178-6. [DOI] [PubMed] [Google Scholar]
- 47.Kammerer CM, Schneider JL, Cole SA, Hixson JE, Samollow PB, O’Connell JR, Perez R, Dyer TD, Almasy L, Blangero J, Bauer RL, Mitchell BD. Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res. 2003;18:2245–2252. doi: 10.1359/jbmr.2003.18.12.2245. [DOI] [PubMed] [Google Scholar]
- 48.Xiong DH, Liu YZ, Liu PY, Zhao LJ, Deng HW. Association analysis of estrogen receptor alpha gene polymorphisms with cross-sectional geometry of the femoral neck in Caucasian nuclear families. Osteoporos Int. 2005;16:2113–2122. doi: 10.1007/s00198-005-2011-4. [DOI] [PubMed] [Google Scholar]
- 49.Ezura Y, Emi M. Genetic analysis of osteoporosis susceptibility: future perspectives on genome wide analysis. Clin Calcium. 2005;15:762–767. [PubMed] [Google Scholar]
- 50.Roef G, Lapauw B, Goemaere S, Zmierczak H, Fiers T, Kaufman JM, Taes Y. Thyroid hormone status within the physiological range affects bone mass and density in healthy men at the age of peak bone mass. Eur J Endocrinol. 2011;164:1027–1034. doi: 10.1530/EJE-10-1113. [DOI] [PubMed] [Google Scholar]
- 51.Gogakos AI, Duncan Bassett JH, Williams GR. Thyroid and bone. Arch Biochem Biophys. 2010;503:129–136. doi: 10.1016/j.abb.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 52.Ris-Stalpers C, Bikker H. Genetics and phenomics of hypothyroidism and goiter due to TPO mutations. Mol Cell Endocrinol. 2010;322:38–43. doi: 10.1016/j.mce.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Ioannidis JP, Ng MY, Sham PC, Zintzaras E, Lewis CM, Deng HW, Econs MJ, Karasik D, Devoto M, Kammerer CM, Spector T, Andrew T, Cupples LA, Duncan EL, Foroud T, Kiel DP, Koller D, Langdahl B, Mitchell BD, Peacock M, Recker R, Shen H, Sol-Church K, Spotila LD, Uitterlinden AG, Wilson SG, Kung AW, Ralston SH. Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J Bone Miner Res. 2007;22:173–183. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Streeten EA, McBride DJ, Pollin TI, Ryan K, Shapiro J, Ott S, Mitchell BD, Shuldiner AR, O’Connell JR. Quantitative trait loci for BMD identified by autosome-wide linkage scan to chromosomes 7q and 21q in men from the Amish Family Osteoporosis Study. J Bone Miner Res. 2006;21:1433–1442. doi: 10.1359/jbmr.060602. [DOI] [PubMed] [Google Scholar]
- 55.Pan F, Xiao P, Guo Y, Liu YJ, Deng HY, Recker RR, Deng HW. Chromosomal regions 22q13 and 3p25 may harbor quantitative trait loci influencing both age at menarche and bone mineral density. Hum Genet. 2008;123:419–427. doi: 10.1007/s00439-008-0490-z. [DOI] [PubMed] [Google Scholar]
- 56.Xiao P, Shen H, Guo YF, Xiong DH, Liu YZ, Liu YJ, Zhao LJ, Long JR, Guo Y, Recker RR, Deng HW. Genomic regions identified for BMD in a large sample including epistatic interactions and gender-specific effects. J Bone Miner Res. 2006;21:1536–1544. doi: 10.1359/jbmr.060717. [DOI] [PubMed] [Google Scholar]
- 57.Pedersen ME, Fortunati D, Nielsen M, Brorson SH, Lekva T, Nissen-Meyer LS, Gautvik VT, Shahdadfar A, Gautvik KM, Jemtland R. Calmodulin-dependent kinase 1beta is expressed in the epiphyseal growth plate and regulates proliferation of mouse calvarial osteoblasts in vitro. Bone. 2008;43:700–707. doi: 10.1016/j.bone.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 58.Ang ES, Zhang P, Steer JH, Tan JW, Yip K, Zheng MH, Joyce DA, Xu J. Calcium/calmodulin-dependent kinase activity is required for efficient induction of osteoclast differentiation and bone resorption by receptor activator of nuclear factor kappa B ligand (RANKL) J Cell Physiol. 2007;212:787–795. doi: 10.1002/jcp.21076. [DOI] [PubMed] [Google Scholar]
- 59.Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 60.Zingg HH, Laporte SA. The oxytocin receptor. TEM. 2003;14:222–227. doi: 10.1016/s1043-2760(03)00080-8. [DOI] [PubMed] [Google Scholar]
- 61.Copland JA, Ives KL, Simmons DJ, Soloff MS. Functional oxytocin receptors discovered in human osteoblasts. Endocrinology. 1999;140:4371–4374. doi: 10.1210/endo.140.9.7130. [DOI] [PubMed] [Google Scholar]
- 62.Colucci S, Colaianni G, Mori G, Grano M, Zallone A. Human osteoclasts express oxytocin receptor. Biochemical and Biophysical Research Communications. 2002;297:442–445. doi: 10.1016/s0006-291x(02)02009-0. [DOI] [PubMed] [Google Scholar]
- 63.Petersson M, Lagumdzija A, Stark A, Bucht E. Oxytocin stimulates proliferation of human osteoblast-like cells. Peptides. 2002;23:1121–1126. doi: 10.1016/s0196-9781(02)00041-4. [DOI] [PubMed] [Google Scholar]
- 64.Breuil V, Amri EZ, Panaia-Ferrari P, Testa J, Elabd C, bert-Sabonnadiere C, Roux CH, Ailhaud G, Dani C, Carle GF, Euller-Ziegler L. Oxytocin and bone remodelling: Relationships with neuropituitary hormones, bone status and body composition. Joint Bone Spine. 2011 doi: 10.1016/j.jbspin.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Frost HM. On the estrogen-bone relationship and postmenopausal bone loss: A new model. J Bone Miner Res. 1999;14:1473–1477. doi: 10.1359/jbmr.1999.14.9.1473. [DOI] [PubMed] [Google Scholar]
- 66.Kusu N, Laurikkala J, Imanishi M, Usui H, Konishi M, Miyake A, Thesleff I, Itoh N. Sclerostin is a novel secreted osteoclast-derived bone morphogenetic protein antagonist with unique ligand specificity. J Biol Chem. 2003;278:24113–24117. doi: 10.1074/jbc.M301716200. [DOI] [PubMed] [Google Scholar]
- 67.Ott SM. Sclerostin and Wnt signaling--the pathway to bone strength. J Clin Endocrinol Metab. 2005;90:6741–6743. doi: 10.1210/jc.2005-2370. [DOI] [PubMed] [Google Scholar]
- 68.Brunkow ME, Gardner JC, Van NJ, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, Alisch RS, Gillett L, Colbert T, Tacconi P, Galas D, Hamersma H, Beighton P, Mulligan J. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]