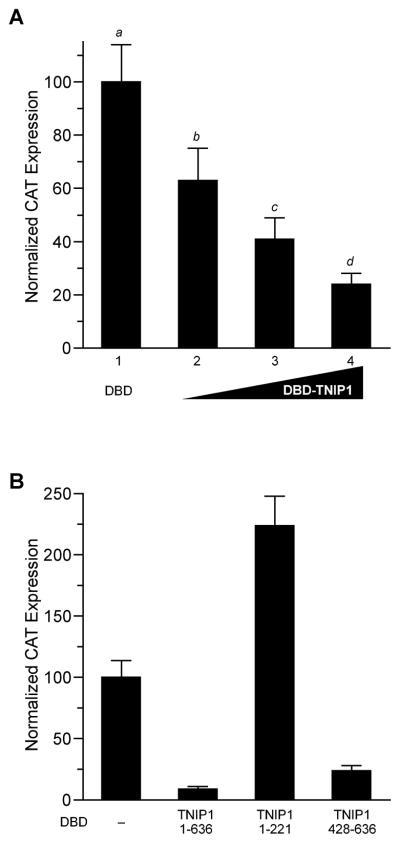
Fig. 6. TNIP1 transcriptional effects in one-hybrid assays.
(A) Full-length TNIP1 exhibits intrinsic transcriptional repression. COS-7 cells transfected with a DBD-regulated reporter, pGal4UAS-tk-CAT, and pGal4DBD with no insert (DBD, first bar) or with increasing amounts, 0.75, 1.50, 3.0μg of pGal4DBD-TNIP1 containing the full-length TNIP1 cDNA in-frame after the DBD coding region (bars 2 – 4, respectively) per well of 6-well plate. pGal4DBD with no insert was included as needed to maintain equal copy number of total DBD constructs for each transfection. CAT expression from pGal4DBD with no insert set to 100%. All values significantly different (a – d) from each other at p < 0.05 or less; 0.75 μg DBD-TNIP1 vs DBD, p < 0.01; higher DBD-TNIP1 amounts against DBD control, p < 0.001 (one-way ANOVA, Newman-Keuls post-hoc test).
(B) Expression in COS-7 cells of the constitutively active pGal4UAS-tk-CAT reporter in response to cotransfection with equal copy number of the pGal4DBD vector (DBD) with no insert (–), TNIP1 full-length (amino acids 1 – 636), or TNIP1 truncations as indicated by amino acid positions in graph.