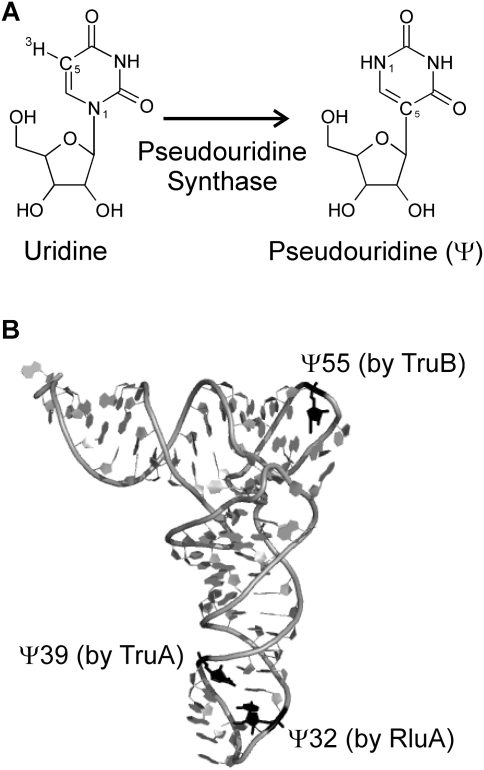
FIGURE 1.
Structure of pseudouridine and modification sites in E. coli tRNAPhe. (A) Uridines in RNA are isomerized to pseudouridines (Ψ) by enzymes called pseudouridine synthases. The N1 and C5 atoms are indicated, which are part of the glycosidic bond in uridines and pseudouridine, respectively. Also, the tritium label at C5 is shown, which is released upon pseudouridine formation in the tritium release assay. (B) The positions of the three pseudouridines found in E. coli tRNAPhe are depicted in black, and the responsible pseudouridine synthase (RluA, TruA, or TruB) is indicated for each modification site. Note that the structure of yeast tRNAPhe shown here does not contain a pseudouridine at position 32 (Protein Data Bank ID code 4TRA [Westhof et al. 1988]). This figure was generated with Pymol (www.pymol.org).