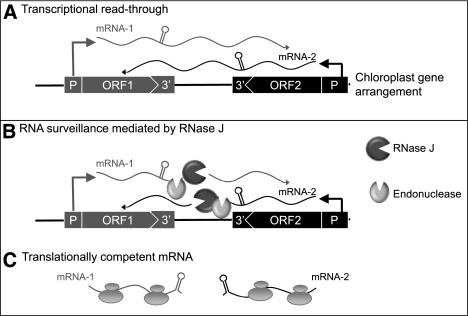
FIGURE 9.
Model for asRNA surveillance by chloroplast RNase J. (A) 3′ UTRs of chloroplast genes inefficiently terminate transcription, resulting in read-through (mRNA-1 and mRNA-2). Where genes are convergently transcribed, even at a distance, asRNA may be synthesized. (B) These pre-mRNAs are first processed by an endonuclease, which could possibly be RNase J itself or another unidentified endonuclease. This creates substrates for the 5′ → 3′ exonucleolytic activity of RNase J. (C) By removing asRNA, RNase J allows accumulation of single-stranded sense RNA that is translationally competent.