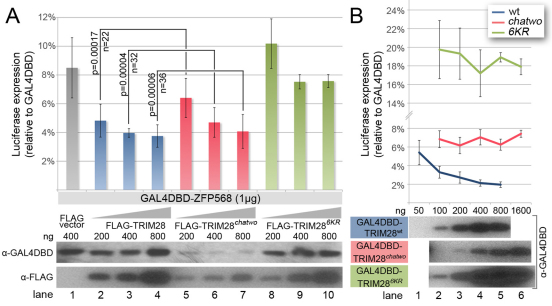
Fig. 6.
chatwo mutations disrupt TRIM28 stability and repressive activity. Luciferase expression from a 5xUAS-luciferase reporter in HEK293T cells, (A) in the presence of GAL4DBD-ZFP568 and either FLAG empty vector (gray bar), increasing amounts of FLAG-TRIM28 (blue), Flag-TRIM28chatwo (red) or Flag-TRIM286KR (green); (B) in the presence of GAL4DBD-TRIM28wt (blue line), GAL4DBD-TRIM28chatwo (red line) or GAL4DBD-TRIM286KR (green line). Note that the dose-dependent effect of GAL4DBD-TRIM28wt (blue line) is lost in GAL4DBD-TRIM28chatwo and GAL4DBD-TRIM286KR mutants (red and green lines, respectively). Luciferase expression is plotted as the percentage relative to GAL4DBD empty vector (100%). Error bars represent s.d. Results in A represent one of nine experiments showing similar results. P-values were calculated using data from all nine experiments (n=total number of data points). Western blots show levels of chimeric proteins normalized for transfection efficiency. Levels of GAL4DBD-ZFP568 in the presence of FLAG-TRIM28chatwo were significantly lower compared with FLAG-vector conditions in six out of seven western blot experiments.