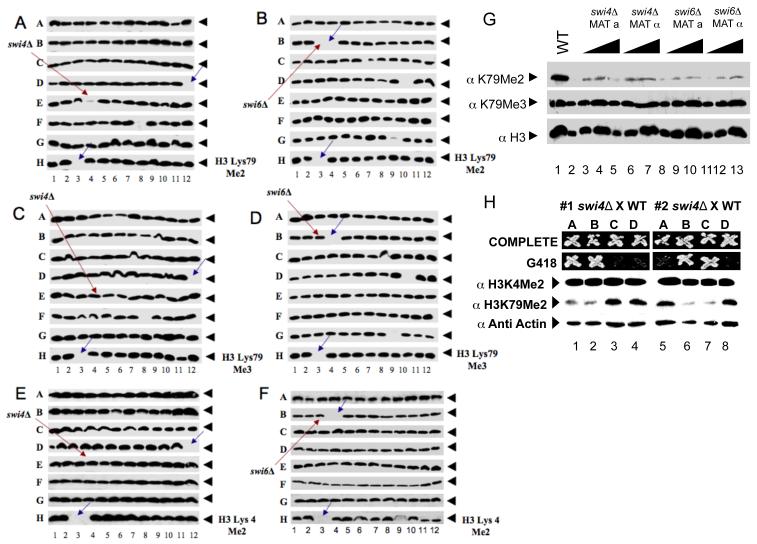
Figure 1. GPS reveals that SWI4 and SWI6 are required for H3K79 di- but not trimethylation.
(A - F) Cell extracts from each of the nonessential yeast gene deletion mutants were subjected to SDS-PAGE, Western blotted, and probed with antibodies specific for (A and B) H3K79 dimethylation or (C and D) trimethylation. (E and F) H3 K4 dimethylation was used as a control. Red arrows represent (A, C, E) swi4Δ and (B, D, F) swi6Δ. Blue arrows indicate empty wells that serve as plate markers. (G) Protein extracts from two different mating strains of swi4Δ and swi6Δ were analyzed as in A-F. Anti-acetyl H3 was probed for a loading control. (H) The swi4::kanMX mutant from the deletion collection was mated with a wild type strain and spores of the resulting tetrads were dissected and scored for the KanR and H3K79 methylation pattern.