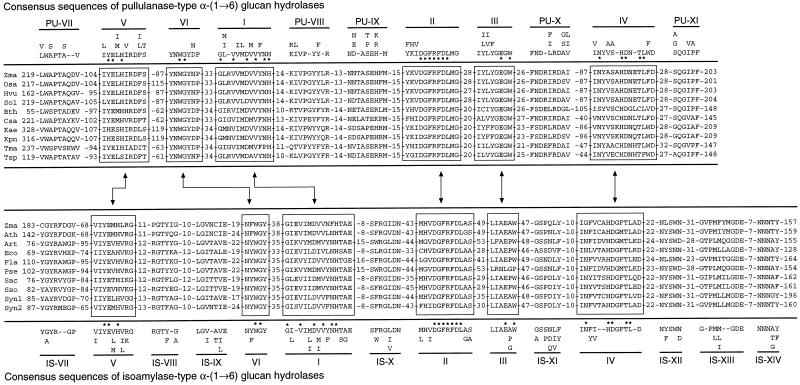
Figure 2.
Multiple sequence alignment of pullulanase- and isoamylase-type DBEs from higher plants and prokaryotes. DBEs are grouped based on characterized enzymatic activity and/or sequence similarity; any polypeptide within a class is significantly more similar to others within that group than to those of the other class. Conservative substitutions in the consensus sequences are noted when they fall into the functional groups defined by Dayhoff and Orcutt (1979), which are AGPST, ILMV, HKR, DENQ, RWY, and C. Residues invariant in all 19 sequences are noted by asterisks. Rare exceptions to the consensus sequence are underlined. Numerals refer to amino acid position beginning at the first ATG codon of the open reading frame. The number of nonconserved amino acids adjacent to each conserved motif is indicated. Conserved motifs in boxes are present in both the pullulanase- and isoamylase-type classes, whereas conserved motifs without boxes are specific to one of the classes as indicated. Motifs I to IV are those defined previously that occur in all members of the α-amylase superfamily (Jesperson et al., 1993), and are numbered accordingly. Abbreviations and accession numbers are provided for the following DBES. Pullulanases and pullulanase-type DBEs: Bth (Bacteroides thetaiotaomicron, U67061); Csa (Caldicellulosiruptor saccharolyticus, L39876); Kae (Klebsiella aerogenes, M16187); Hvu (Hordeum vulgare, AF022725); Kpn (Klebsiella pneumoniae, X52181); Osa (Oryza sativa, D50602); Sol (Spinacia oleracea, X83969); Tma (Thermotoga maritima, AJ001087); Tsp (Thermus sp. IM6501, AF060205); Zma (Z. mays, AF080567, this study). Isoamylases and isoamylase-type DBEs: Art (artificial gene, A10906); Ath (Arabidopsis, AF002109); Eco (E. coli, U18997); Fla (Flavobacterium sp., U90120); Psu (Pseudomonas sp., A28109, A37035); Sac (Sulfolobus acidocaldarius, D83245); Sso (Sulfolobus solfataricus, Y08256); Syn1 (Synechocystis sp., U44761); Syn2 (Synechocystis sp., D90908); and Zma (Z. mays, U18908).