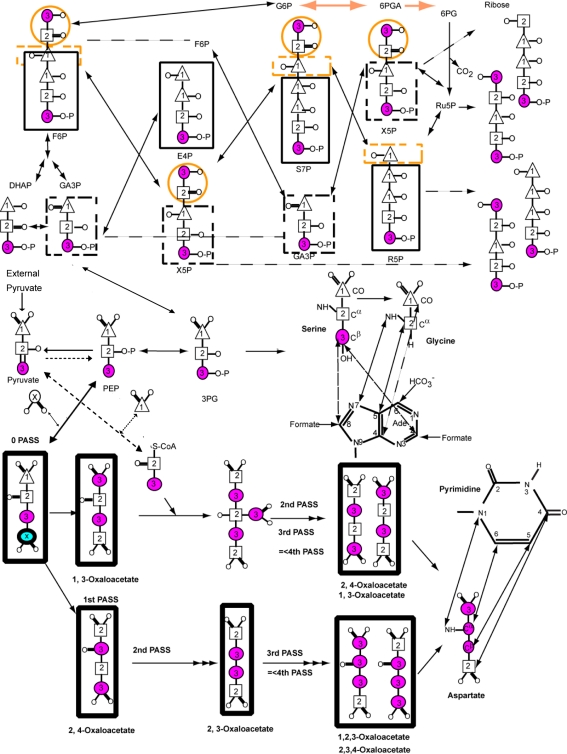
Fig. 1.
Major metabolic pathways involved in the production of nucleic acid nucleotides from pyruvate, including key steps in glycolysis, gluconeogenesis and several passes through the tricarboxylic (TCA) cycle as derived from Covert and Palsson (2002). With the E. coli strain lacking succinate and malate dehydrogenases (DL323), the oxidative branch of the pentose phosphate pathway remains intact but the TCA cycle is severed in two places such that the oxaloacetate is derived exclusively from carboxylation of phosphoenolpyruvate (PEP) and the resulting label is not diluted by the TCA cycle. Atom labels for the terminal (3) carbon (magenta and thin circle), the central (2) carbon (square), and the terminal (1) carbon (triangle) of pyruvate are highlighted. Positions that are enriched due to the presence of 13CO2 in the growth medium are shown with a circled x. Pyrimidine base derived from the oxaloacetate (OAA) produced by carboxylation of PEP is shown via the aspartate intermediate. This OAA cannot be used as a substrate in the first and subsequent rounds of the TCA cycle because of the two mutations. Consequently OAA derived aspartate amino acid can be produced with 13C labeling at only the Cα (C6) position if [2-13C]-pyruvate is used. If [3-13C]-pyruvate is used only Cβ (C5) position is labeled. In either case carboxylation of PEP leads to labeling of the Cγ (C4) position. Similarly the labeling pattern of purines from glycine derived from 3PG are shown such that if [2-13C]-pyruvate is used only the Cα position of Gly and therefore C5 position of the purine ring is labeled. Otherwise if [3-13C]-pyruvate is used the CO of Gly and therefore C4 of purine ring is labeled, and the labeling of the Cβ position of Ser also leads to labeling of the purine C2 and C8 positions