Figure 3. Characteristics of BC-1 and BC-3 PAR-CLIP libraries.
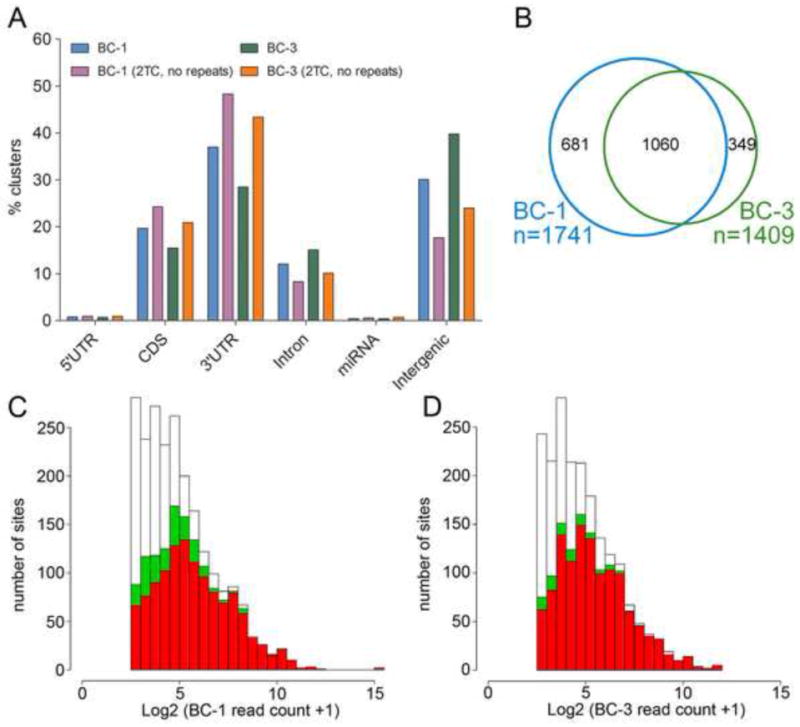
(A) Location of PAR-CLIP clusters within the human genome. “2TC, no repeats”: only clusters not mapping to repeats and with T to C conversions at ≥ 2 locations were analyzed. (B) Venn diagram showing the overlap of mRNAs with 3′UTR sites for KSHV miRNAs between BC-1 and BC-3 libraries. Only KSHV miRNAs expressed in both cell lines were considered. (C) Read depth for KSHV miRNA targets sites found in BC-1 (white), read depth for the subset of these clusters that were also recovered in BC-3 with ≥ 1 (green) or ≥2 (red) locations with T to C conversions. (D) as in (C), except that sites from BC-3 are in white and the sites that are also recovered in BC-1 are in green and red. See also Tables S5, 6.
